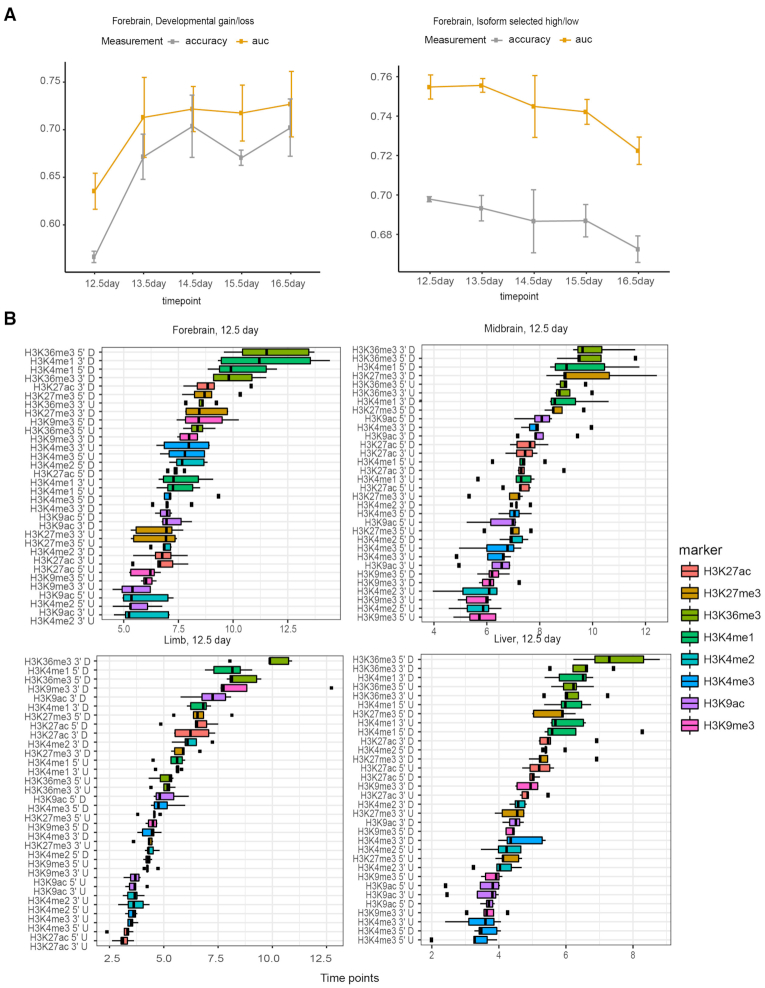
Figure 3.
Model performance and important histone modifications associated with skipped exon selection in different tissues and timepoints (A) Accuracy and AUC values of random forest models built based on hPTM signals on the flanking regions of skipped exons in forebrain across developmental time points. (B) H3K36me3 and H3K4me1 are the most predictive hPTM in differentiating skipped exon inclusion categories. Boxplot of important score generated by random forest model in different tissues at E12.5 shows several types of hPTM are key predictors. Importance score is calculated based on 5-fold cross validation.