Figure 1.
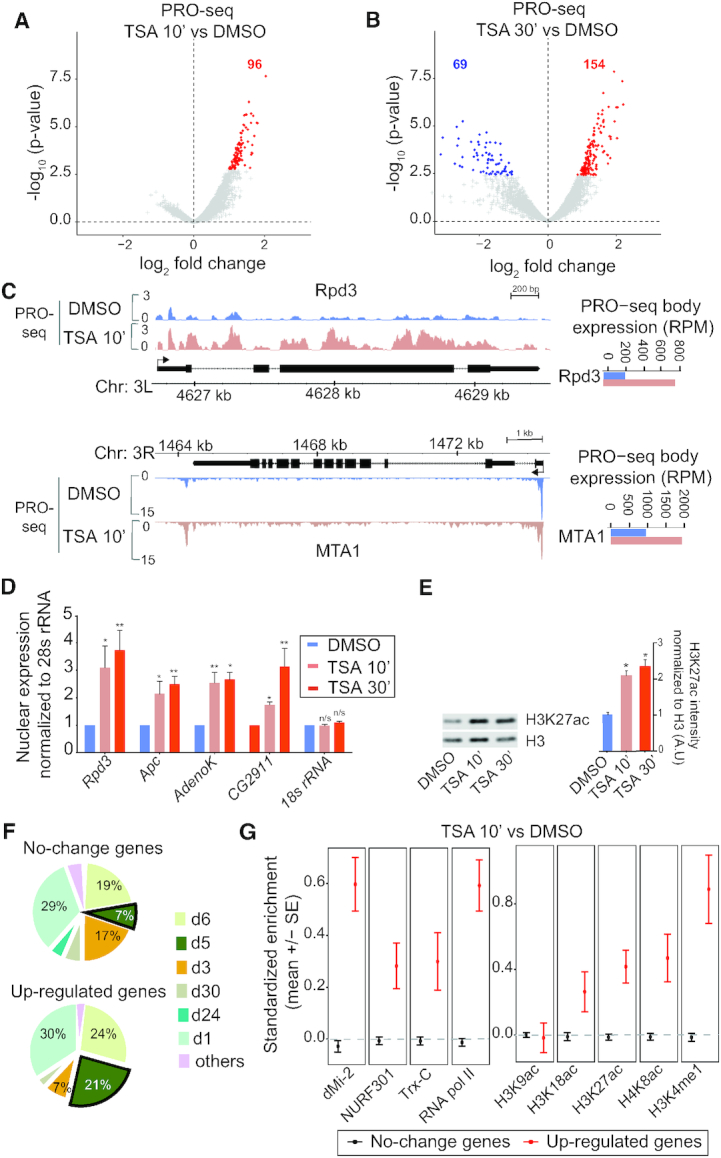
Histone deacetylase inhibition results in rapid transcriptional up-regulation of genes associated with a distinct chromatin state. (A and B) PRO-seq gene body (500 bp from TSS – 100 bp upstream of TES) log2 fold change (x-axis) versus -log10P-values (y-axis) in Drosophila S2 cells treated with the HDAC inhibitor TSA versus DMSO control for 10 min (A) or 30 min (B). (C) PRO-seq signal in DMSO control and TSA-treated S2 cells shown for the Rpd3 and MTA1 loci. (D) RT-qPCR with nuclear RNA isolated from S2 cells treated with DMSO or TSA for 10 or 30 min normalized against 28S rRNA. n = 4. Error bars represent SEM and significant differences between control and TSA-treated cells (two-tailed paired t-test) are indicated by asterisks, * P< 0.05, ** P< 0.01. (E) Western blot showing H3K27 acetylation and total H3 in DMSO and TSA-treated cells. The H3K27ac signal was normalized to total H3 and error bars represent SEM, n = 4. Significant differences between control and TSA-treated cells (two-tailed paired t-test) are indicated by asterisks, * P< 0.05. (F) Association of promoter regions (±500 bp around TSS) of 96 up-regulated genes within 10 min of TSA and genes that do not change their expression with modENCODE chromatin states. (G) Average standardized enrichment scores (z-scores) of dMi-2 (NuRD complex), NURF301 (NURF complex), Trithorax (Trx-C), Pol II and histone modifications in unchanged and 96 up-regulated gene promoters.
