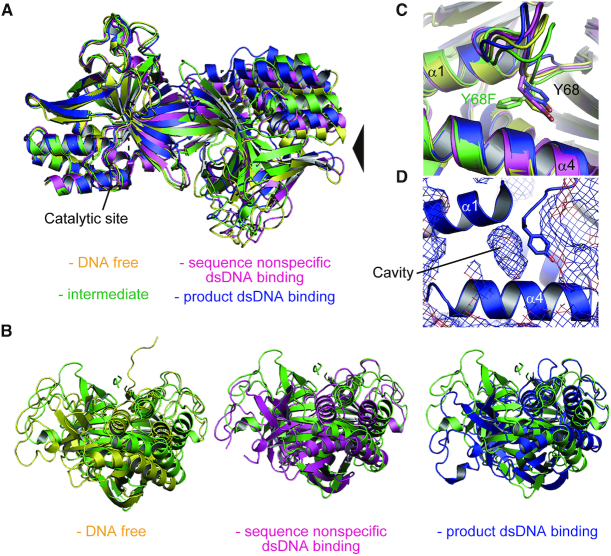
Figure 3.
Structural comparison of the R.PabI dimers. (A) Superposition of the R.PabI structures in the DNA-free state (PDB ID: 2DVY, yellow), the sequence-nonspecific dsDNA-binding state (PDB ID: 5IFF, magenta), the intermediate state (green) and the product dsDNA-binding state (PDB ID: 3WAZ, blue). The structures were superposed using the coordinates of chain A structures. The positions of the catalytic sites are indicated by black dotted squares. (B) Side views of (A). The viewpoint is indicated by a black triangle in (A). (C) Side chain flipping of Y68F observed in the intermediate state. The side chains of Y68 and Y68F are shown by stick models. (D) Cavity adjacent to Y68. The protein surface of R.PabI (PDB ID: 3WAZ) is shown in mesh.