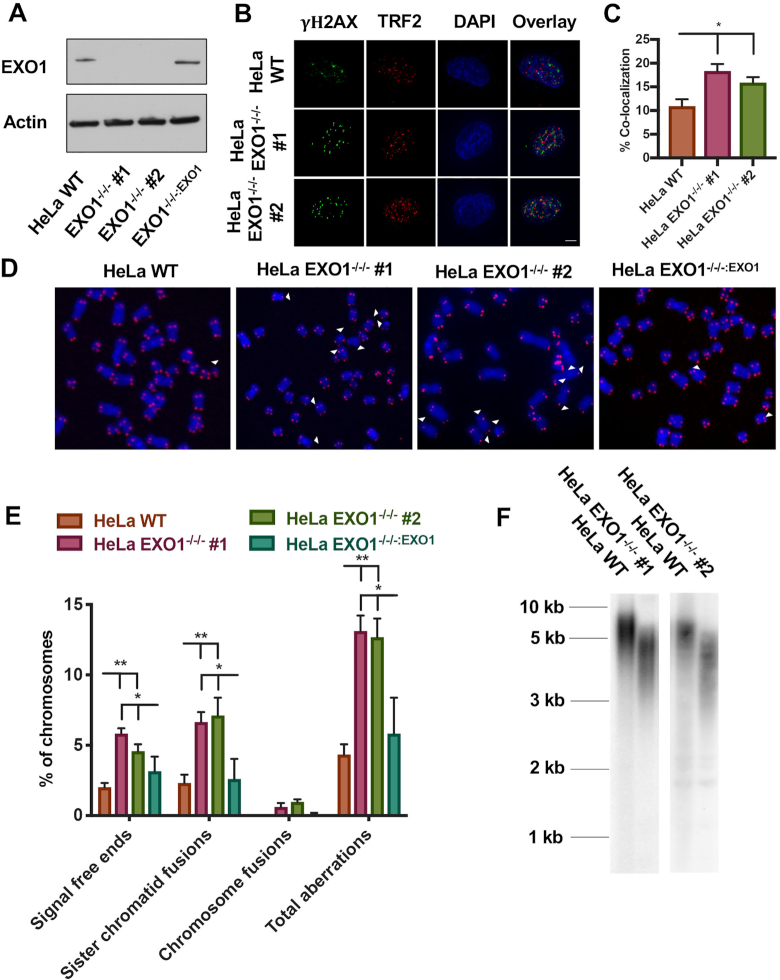
Figure 1.
Generation and characterization of telomere defects in EXO1-null HeLa cells. (A) Western blot confirmation of EXO1 expression in the parental wild-type (WT) HeLa cells, as well as two EXO1-null clones (#1 and #2) and EXO1−/−/−:EXO1, an EXO1-null clone complemented with an EXO1 cDNA. Actin was used as a loading control. (B) DNA damage assessment at telomeres with a TIF analysis. TRF2 was used as a telomere marker and gamma-H2AX as a marker of DNA damage. Scale bar = 10 μm. (C) Quantification of the TIF analysis from (B). A paired t-test was used to evaluate statistical significance; n = 150 from three independent experiments. (D) Representative images of T-FISH analyses of EXO1-HeLa cell lines. Chromosomes were DAPI stained (blue) and telomeres imaged with a PNA probe (red). Abnormalities are marked with white arrowheads. (E) Quantification of T-FISH from (D). Sister chromatid fusions, chromosome:chromosome fusions and signal-free ends were scored. A one-way ANOVA analysis followed by Tukey's post-hoc test was used to determine statistical significance; n = 6000 from three independent experiments. (F) Telomere restriction fragment (TRF) assessment of telomere length in HeLa EXO1-null cell lines.