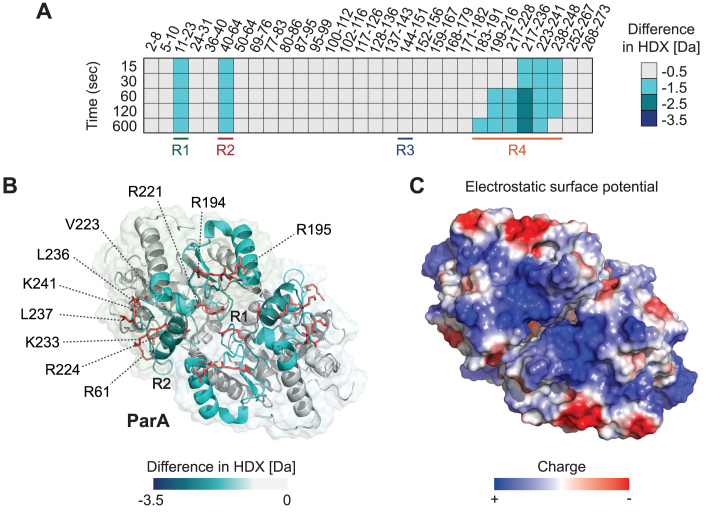
Figure 5.
Hydrogen/deuterium exchange (HDX) analysis of the C. crescentus ParA–DNA interaction. (A) ParA was incubated in deuterated buffer containing 1 mM ATPγS in the absence or presence of a 14 bp dsDNA oligonucleotide (ran14-up/ran14-lo). The heat plot shows the maximal differences in deuterium uptake between the DNA·ParA complex and MipZ alone at different incubation times for a series of representatives peptides (see Dataset S2 for the full list of peptides). The color code is given on the right. All experiments were performed in the presence of ATPγS to lock the protein in the dimeric state. The four different regions that are protected upon DNA binding to MipZ (see Figure 4) or ParA are indicated at the bottom. (B) Mapping of the maximum differences in HDX observed upon DNA binding onto a structural model of ParA, generated with HpParA (PDB ID: 6IUB) (39) as a template. A surface representation of the structure is shown in the background. For clarity, the ATP and Mg2+ ligands are not shown. (C) Electrostatic surface potential of the ParA dimer. The color code is given at the bottom.