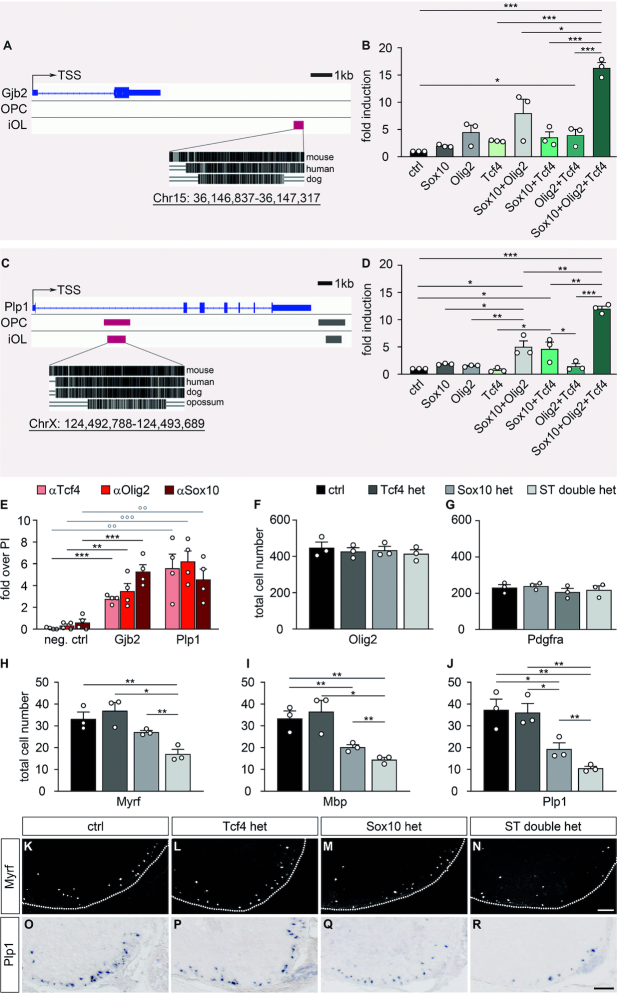
Figure 10.
Joint influence of Tcf4, Olig2 and Sox10 on oligodendroglial gene expression. (A, C) Localization of evolutionary conserved regulatory regions with Olig2 binding capacity (red box) in OPCs or immature oligodendrocytes (iOL) in the Gjb2 (A) and Plp1 (C) genomic region relative to transcriptional start site (TSS) and exons (blue boxes). The exact genomic position according to rn4 is given at the bottom. (B, D) Activation of luciferase reporters under control of these regulatory regions from the Gjb2 (B) and Plp1 (D) genes in transiently transfected N2a cells by Olig2, Tcf4L, Sox10 and combinations thereof as indicated below the bars (n = 3; presented as fold inductions ± SEM, transfections without added transcription factors set to 1 for each regulatory region). (E) Chromatin immunoprecipitation on cultured differentiating oligodendroglial cells with antibodies directed against Tcf4, Olig2 and Sox10. Enrichment of Gjb2, Plp1 and control (neg. ctrl) genomic region in the immunoprecipitates was determined relative to preimmune/IgG controls (PI) that were arbitrarily set to 1. Experiments were performed four times with each PCR in triplicate. (F–J) Quantification of the number ± SEM of Olig2-positive oligodendroglial cells (F), Pdgfra-positive OPCs (G), Myrf-, Mbp- or Plp1-positive oligodendrocytes (H–J) per thoracic spinal cord section in wildtype control (ctrl), Tcf4 het, Sox10 het and ST double het mice at E18.5 (n = 3 mice for each genotype, counting three separate sections). (K–R) Immunohistochemical stainings with antibodies directed against Myrf (K-N) and in situ hybridizations with a probe directed against Plp1 (O–R) on spinal cord sections from wildtype (ctrl; K, O), Tcf4 het (L, P), Sox10 het (M, Q) and ST double het (N, R) mice at E18.5. The right ventral horn is shown. For immunohistochemical stainings, the spinal cord was placed on a black background and demarcated by a stippled line. Scale bar: 100 μm. Statistical significance was determined by one way ANOVA with Bonferroni correction (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).