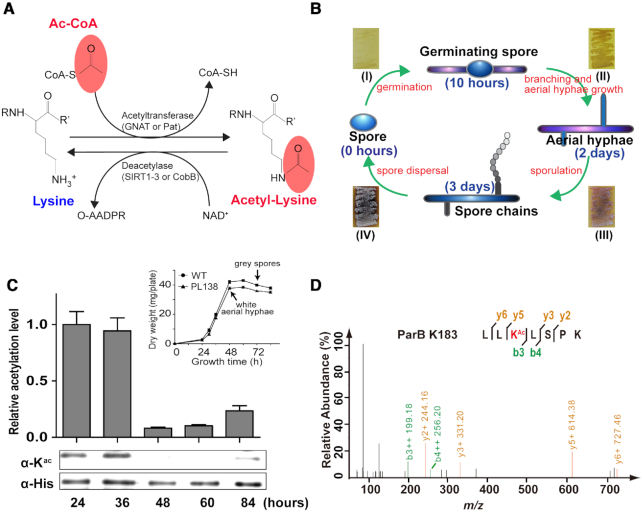
Figure 1.
Identification of the chromosome partitioning protein ParB as an acetylation target in S. coelicolor. (A) Chemical structures of a lysine residue and an acetylated lysine. The corresponding substrates and the products of acetylation or deacetylation are shown. (B) Schematic representation of the S. coelicolor life cycle. S. coelicolor vegetative hyphae germinate from a single spore. They branch and develop into aerial filaments based on nutrient availability and other environmental signals. Aerial hyphae develop after ∼2 days of culture and then turn into spore chains. The life cycle restarts after the spores are mature and dispersed. Corresponding phenotypes (I–IV) of S. coelicolor on MS solid medium are shown as an example. (C) Analysis of the ParB acetylation level throughout the S. coelicolor cell cycle on MS solid culture. The His-tagged ParB proteins were isolated from the S. coelicolor PL138 strain at different time points (24, 36, 48, 60 and 84 h) and then quantified by western blotting. Their relative acetylation levels were normalized to the loading protein levels (see Materials and Methods). The results show that the ParB acetylation level was decreased after 48-h cultivation, which is at a growth phase that cultures are composed of both vegetative hyphae and white aerial hyphae (corresponding to growth phenotype III in Figure 1B). Similar growth curves were monitored between PL138 and wild-type S. coelicolor (upper right panel), ruling out possible impacts of the labelled ParB on cell growth. One representative result of triplicate measurements is shown. HPLC/MS-MS analysis identified Lys-183 (D) as one of the two ParB acetylation sites. The ParB protein was isolated from S. coelicolor after a 36-h cultivation and then digested and subjected to HPLC/MS-MS analyses.