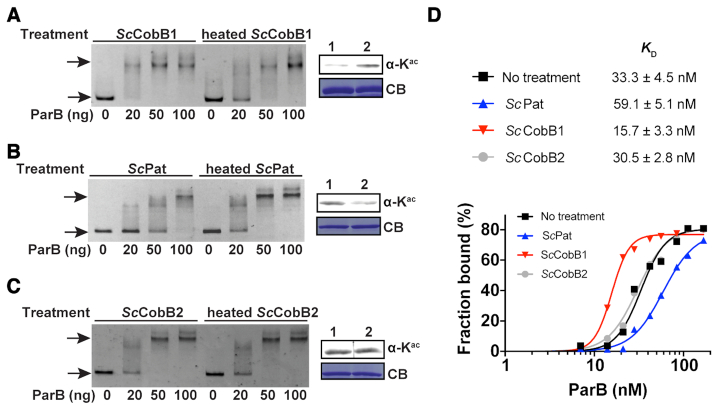
Figure 3.
Deacetylation enhances the DNA-binding affinity of ParB in vitro. (A) An EMSA was performed by incubating a FAM-labelled parS sequence with varying amounts of ParB proteins that were pre-treated with active or heat-inactivated ScCobB1. Free DNA and DNA–protein complexes (indicated by arrows) were separated by 5% polyacrylamide gel electrophoresis. Compared to heated-ScCobB1-treated ParB, 20 ng (28 nM) deacetylated ParB can efficiently bind to parS fragments. The ParB acetylation level induced by active ScCobB1 (lane 1) or heated ScCobB1 (lane 2) was confirmed by western blot (right panel). (B) EMSA results of ParB that was pre-treated with active or heat-inactivated ScPat. (C) EMSA results of ParB treated with active or heat-inactivated ScCobB2. (D) Quantification of the DNA-binding affinity of ParB treated with ScCobB1, ScPat, or ScCobB2. The percentages of DNA bound [bound / (bound + free DNA)] in each reaction were plotted versus the concentrations of ParB (see Supplementary Figure S3 for details). The binding constants KD were determined by fitting a non-linear regression with the method of specific binding with a Hill slope.