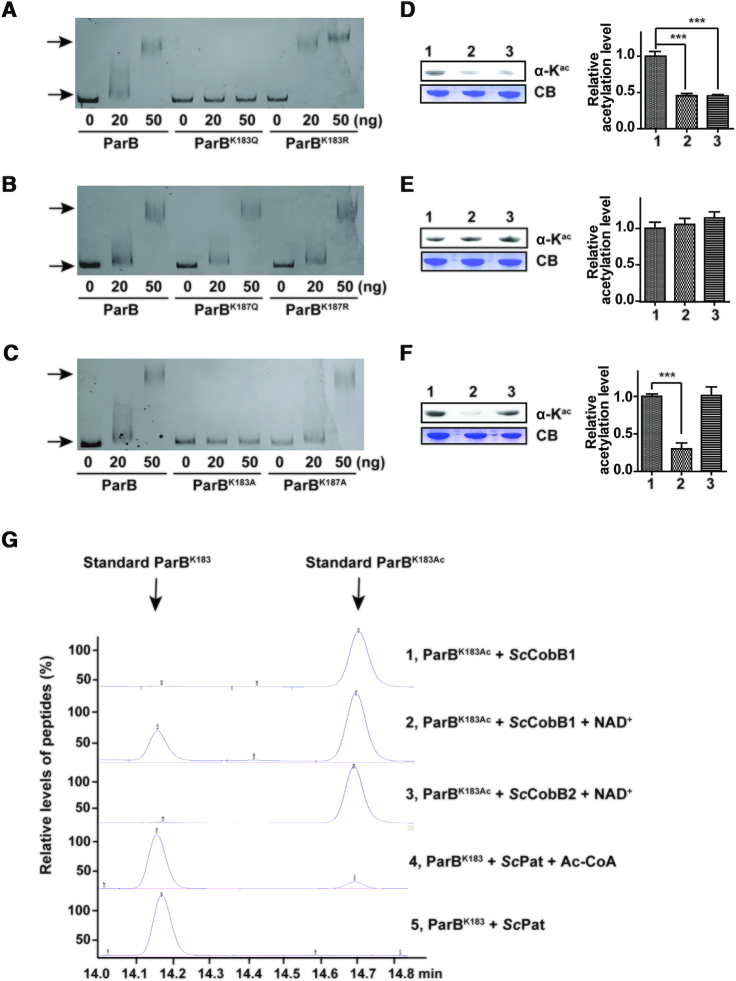
Figure 4.
The Lys-183 residue is the key acetylation site in ParB. (A) Mutation at Lys-183 alters the ParB DNA-binding affinity. An EMSA was performed using purified ParB, ParBK183Q, and ParBK183R as described in Figure 3. (B) The DNA-binding affinity of ParB was unaffected when Lys-187 was mutated. An EMSA was performed using purified ParB, ParBK187Q, and ParBK187R. (C) A lysine to alanine mutation (K to A) confirmed that Lys-183 but not Lys-187 alters the ParB DNA-binding affinity. An EMSA was completed using purified ParB, ParBK183A, and ParBK187A. The acetylation levels of ParB variants were determined by western blotting using a pan anti-acetyllysine antibody. The protein samples analysed in (D), (E) and (F) correspond to the ParB variants in (A), (B) and (C), respectively. One representative result of triplicate measurements is shown. The relative acetylation levels of ParB variants were quantified by ImageJ and normalized against their protein levels (right panels). (G) The regulation of ParB acetylation at Lys-183 was confirmed by HPLC. ParB peptides with an acetylated or non-acetylated Lys-183 residue were synthesized and incubated with ScPat, ScCobB1, or ScCobB2 at 30°C for 2 h. Reactions were terminated by the addition of 1% TFA, after which the samples were analysed by HPLC. Arrows indicate the retention time of the standard acetylated and non-acetylated peptides.