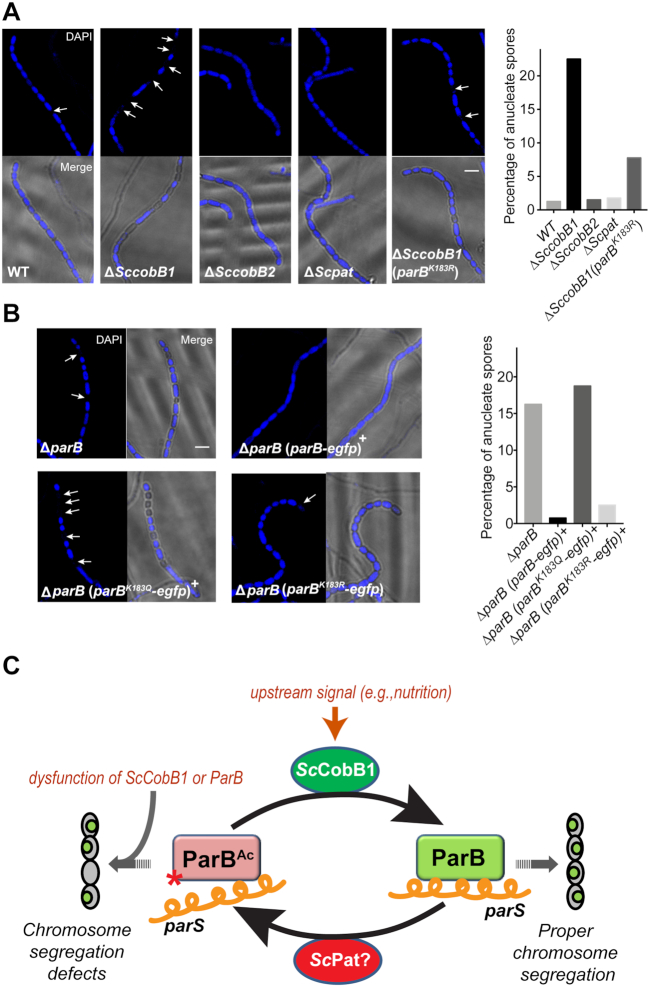
Figure 7.
Exorbitant acetylation of ParB leads to defective cell sporulation in S. coelicolor. Cells were harvested after a 96-h cultivation on MS solid medium to analyse their sporulation by fluorescence microscopy. DAPI staining of the chromosome revealed a higher percentage of anucleate spores (indicated with arrows) in ΔSccobB1 (A), ΔparB (B), and the acetylation mimicking ParB-producing (ParBK183Q) strain (B) than those in wild-type S. coelicolor. Moreover, the sporulation defects in ΔparB and ΔSccobB1 were fully (B) or partially (A) recovered by the deacetylation mimicking ParB (ParBK183R), respectively. The proportions of anucleate spores are shown in the right panels (400 spores calculated each). All scale bars, 2 μm. A model for the regulation of ParB acetylation in S. coelicolor is proposed in (C). The ParB binding affinity to the centromere-like parS sequence is repressed by acetylation, which is absolved by ScCobB1 deacetylation that triggered by the proper upstream signals (e.g. nutrition depletion). The exorbitant acetylation due to the dysfunction of ScCobB1 impedes the formation of chromosome segregation complexes and may lead to the generation of anucleate spores. Red star, acetylation at Lys-183 of ParB; orange helical line, parS; small green oval, S. coelicolor chromosome.