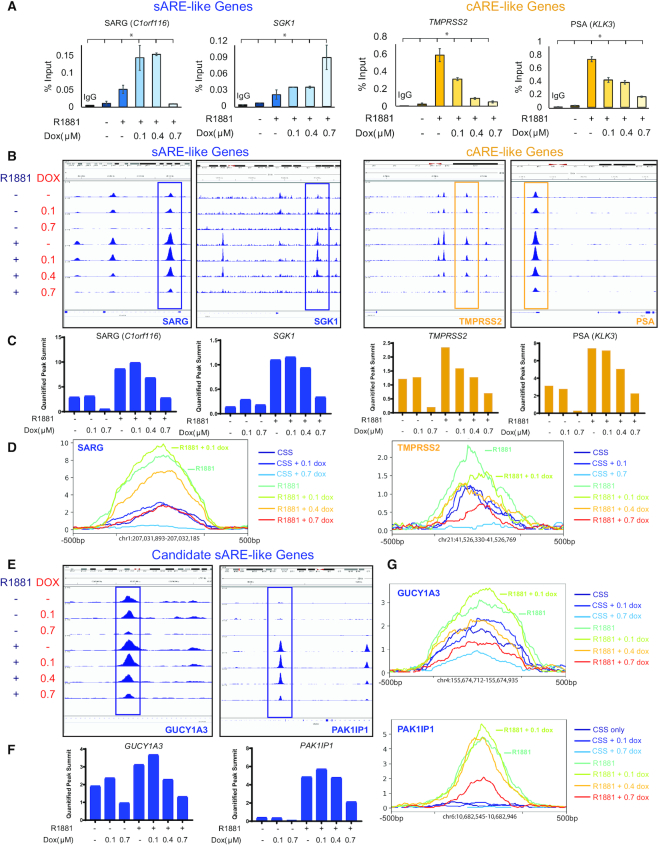
Figure 5.
Dox differentially affects AR recruitment to chromatin, enhancing sARE binding and decreasing cARE binding at low concentrations. (A) ChIP-PCR in LNCaP cells with AR antibodies, IgG control, and purified DNA was quantified by qPCR. Left (blue) – qRT-PCR targeting promoter regions of cARE-driven PSA (KLK3) and TMPRSS2. Right (yellow) - qRT-PCR targeting sAREs in the promoter of SGK1 and intron 1 of SARG (C1orf116) (* indicates P < 0.001). (B) Corresponding ChIP-seq peaks, including those analyzed by qPCR, visualized by Integrative Genomics Viewer (IGV) (60). Peaks that are boxed are enhanced by low-dose 0.1 μM dox in the sARE-like genes of SGK1 and SARG (C1orf116), and show a dose-dependent decrease in the cARE-like genes PSA (KLK3) and TMPRSS2. (C) Read count values for the summits of each peak illustrated by boxes in B graphed for each ChIP condition (quantified peak summits). (D) Isolated peak traces from peaks boxed in part B graphed together on the same axis to illustrate relative area under the curve. (E) ChIP-seq peaks of candidate sARE-like genes identified by RNA-seq, GUCY1A3 and PAK1IP1 visualized by IGV, with sARE-like peaks boxed as in part B. (F) Read count values for the summits of each peak illustrated by boxes in E graphed for each ChIP condition (quantified peak summits). (G) Isolated peak traces from peaks boxed in part E graphed together on the same axis.