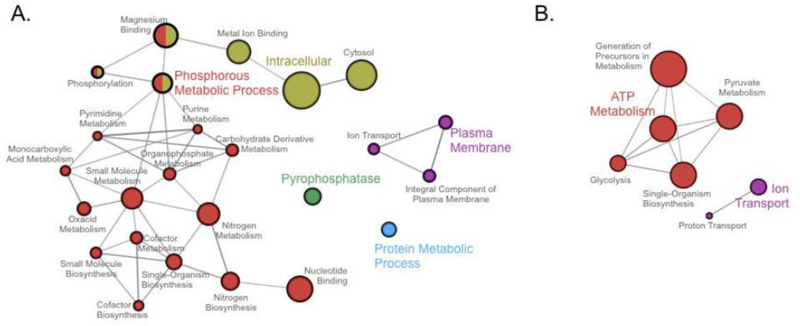
Figure 4:
Global network of cellular processes involved in the resistance of bacteria to fluoride. (A) Conserved molecular functions and cellular components of (A) genes regulated by the fluoride riboswitch, as reported by Weinberg et al. 2010, and (B) altered genes in fluoride resistant bacteria (Zhu et al. 2012; Liao et al. 2015; 2016; Ma et al. 2016; Liu et al. 2017). Genes were converted to their E. coli homologs, and duplicates were discarded. Data was analyzed on Cytoscape using ClueGO. Node size corresponds with the number of genes per category, and similar colored nodes denote a similar cluster in function.