Figure 5. Construction of the multi-reporter DHFR BAC by BAC-MAGIC.
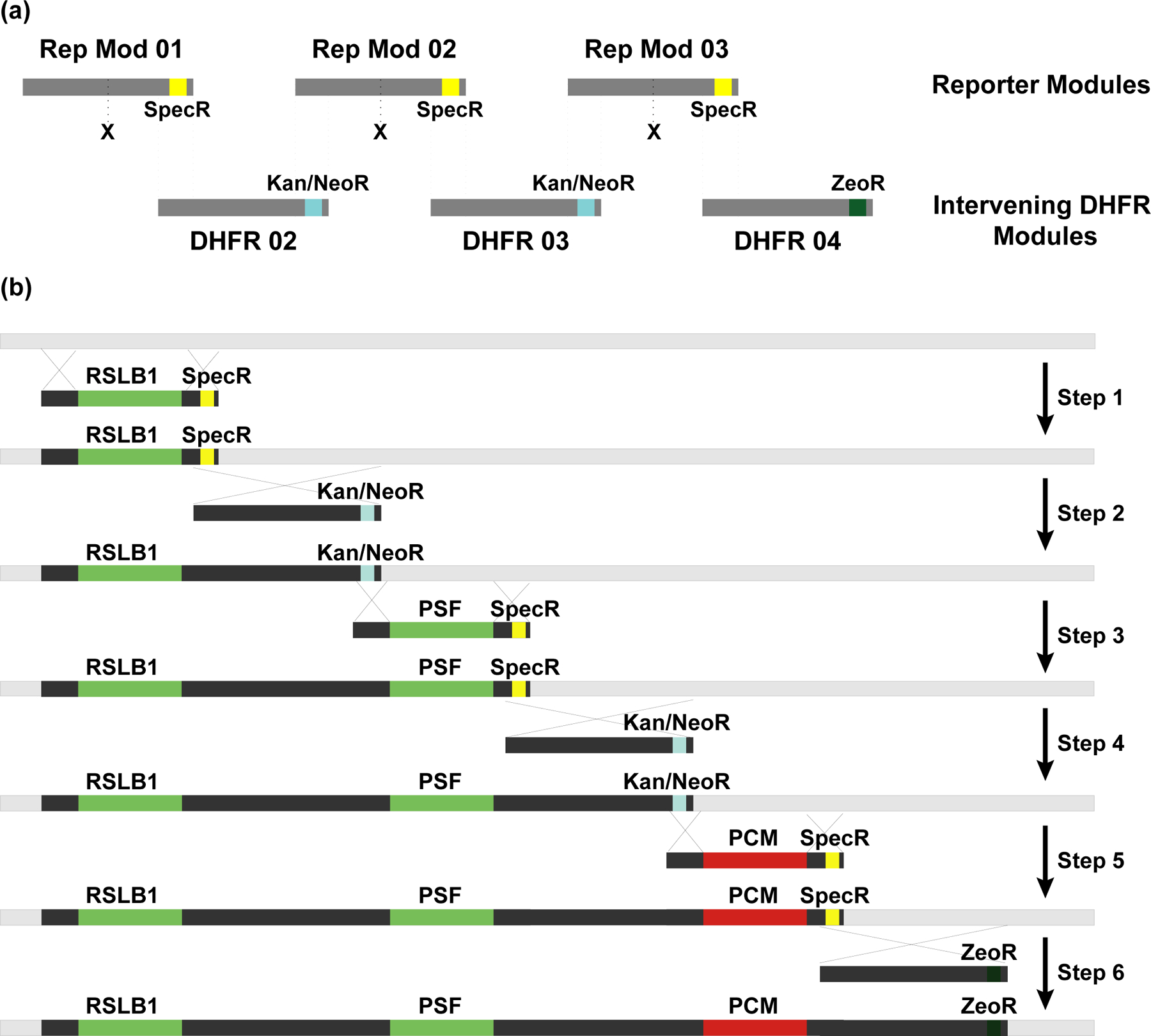
(a) Modular design of BAC-MAGIC. Reporter module 01, 02 and 03 contain reporter gene expression cassettes (X), DHFR BAC homologous sequences (dark gray), and Spectinomycin resistance markers (SpecR, yellow) near the 3’ ends for bacterial selection. Intervening DHFR module 02, 03 and 04 contain DHFR BAC homologous sequences (dark gray), and antibiotic resistance markers near the 3’ ends (Kanamycin/Neomycin resistance marker (Kan/NeoR, blue) in module 02 and 03 for bacterial selection, and Zeocin resistance marker (ZeoR, dark green) in module 04 for dual selection in bacterial or mammalian cells). The dotted lines mark homologous regions between the reporter modules and the intervening DHFR modules. (b) Six sequential steps of BAC recombineering introduce three reporter expression cassettes, RPL32-driven SNAP-tagged Lamin B1 (RSLB1), PPIA-driven SNAP-tagged Fibrillarin (PSF), and PPIA-driven mCherry-Magoh, onto the DHFR BAC (light gray) with ~10 kb of intervening DHFR BAC sequences (dark gray). Homologous regions are indicated by crossed lines.
