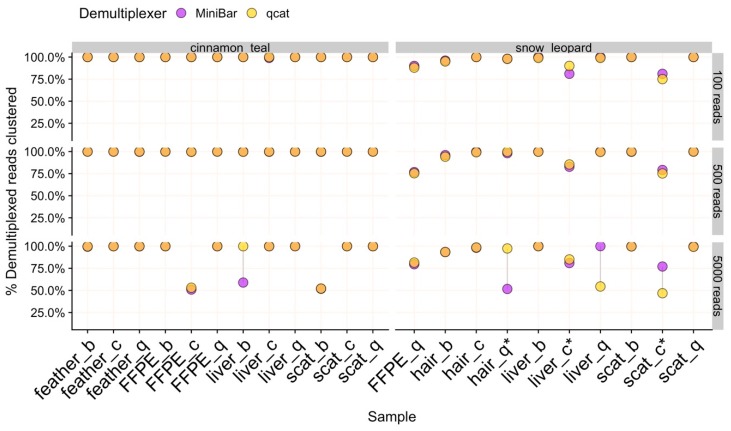
Figure 3.
The percent of demultiplexed reads used to generate the final consensus sequence for 100R, 500R, and 5KR subsets for each species. Samples are labeled by tissue type and extraction method (b = biomeme, c = chelex, q = qiagen). Points are linked by a grey line to show difference in values from demultiplexers. Overlapping areas in orange indicate similar results for Minibar and qcat analyses. Asterisks (*) indicate samples with cinnamon teal contamination.