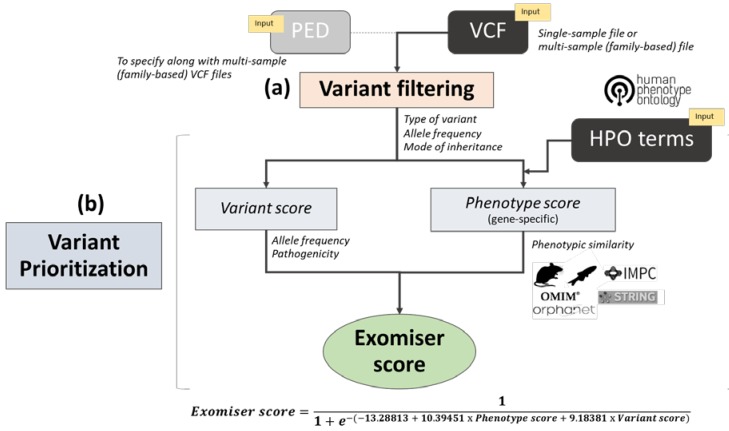
Figure 1.
Overview of the Exomiser workflow analysis: The diagram depicts the two main steps in an Exomiser analysis: (a) variant filtering and (b) variant prioritization. A single-sample variant call format (VCF) file and corresponding list of Human Phenotype Ontology (HPO) terms are mandatory inputs. If a multi-sample VCF file (from a nuclear family) is used in the analysis, the user must provide a corresponding pedigree file. In the filtering step, variants are filtered according to type of variant, allele frequency in selected databases, and mode of inheritance as per user-defined options and values. In the prioritization step, a variant score is calculated based on allele frequency and pathogenicity as predicted by user-defined in silico algorithms, together with a gene-specific phenotype score based on the semantic similarity of the patient’s HPO terms and phenotypic annotation in known human disease gene, mouse, zebrafish and protein–protein interaction databases. Finally, the Exomiser score is obtained from the variant score and phenotypic score within a logistic regression classifier framework and used for variant prioritization. PED: Pedigree, OMIM: Online mendelian inheritance in man, IMPC: International mouse phenotyping consortium, STRING: Search tool for the retrieval of interacting genes/proteins.