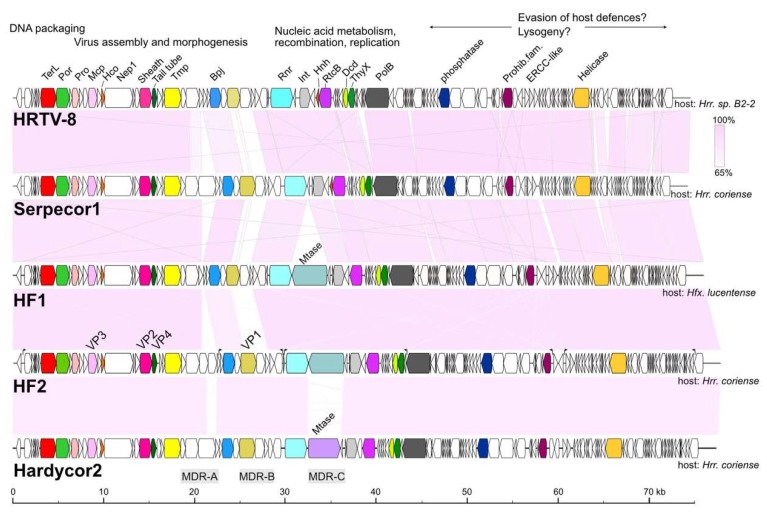
Figure 4.
Genome maps of haloviruses Serpecor1 and Hardycor2 along with their closest relatives, HF1, HF2 and HRTV-8. Pink shading between the maps indicates nucleotide similarity values above 65% (scale at top right). Virus names are given at the left, host species are indicated at the right, and protein names are given at the top of HRTV-8, near the corresponding colored CDS. Corresponding genes on each map have the same color. Scale at bottom represents DNA length in kb. At top are headings indicating the known or likely functional modules (DNA packaging, Virus assembly, etc.). Major Divergent Regions (MDR) are indicated below the Hardycor2 map. Protein names are: TerL, large subunit terminase; Por, portal protein; Pro, prohead protease; Mcp, major capsid protein; Hco, head connector protein; Nep1, neck protein of type 1; Sheath, tail sheath protein; Tail tube, tail tube protein; Tmp, tape measure protein; Bpj, baseplate J family protein; Rnr, ribonucleotide reductase; Int, site-specific integrase; Hnh, HNH-endonuclease; RtcB, tRNA splicing ligase RtcB; Dcd, dCTP deaminase; ThyX, thymidylate synthase; PolB, DNA polymerase elongation subunit (family B); Prohib. fam., prohibitin family protein; ERCC-like, ERCC nuclease family; Helicase, ATP-dependent DNA helicase; Mtase, methyltransferase. The virus structural proteins of HF2 (VP1–VP4) are indicated just above the HF2 genome map.