Fig 5.
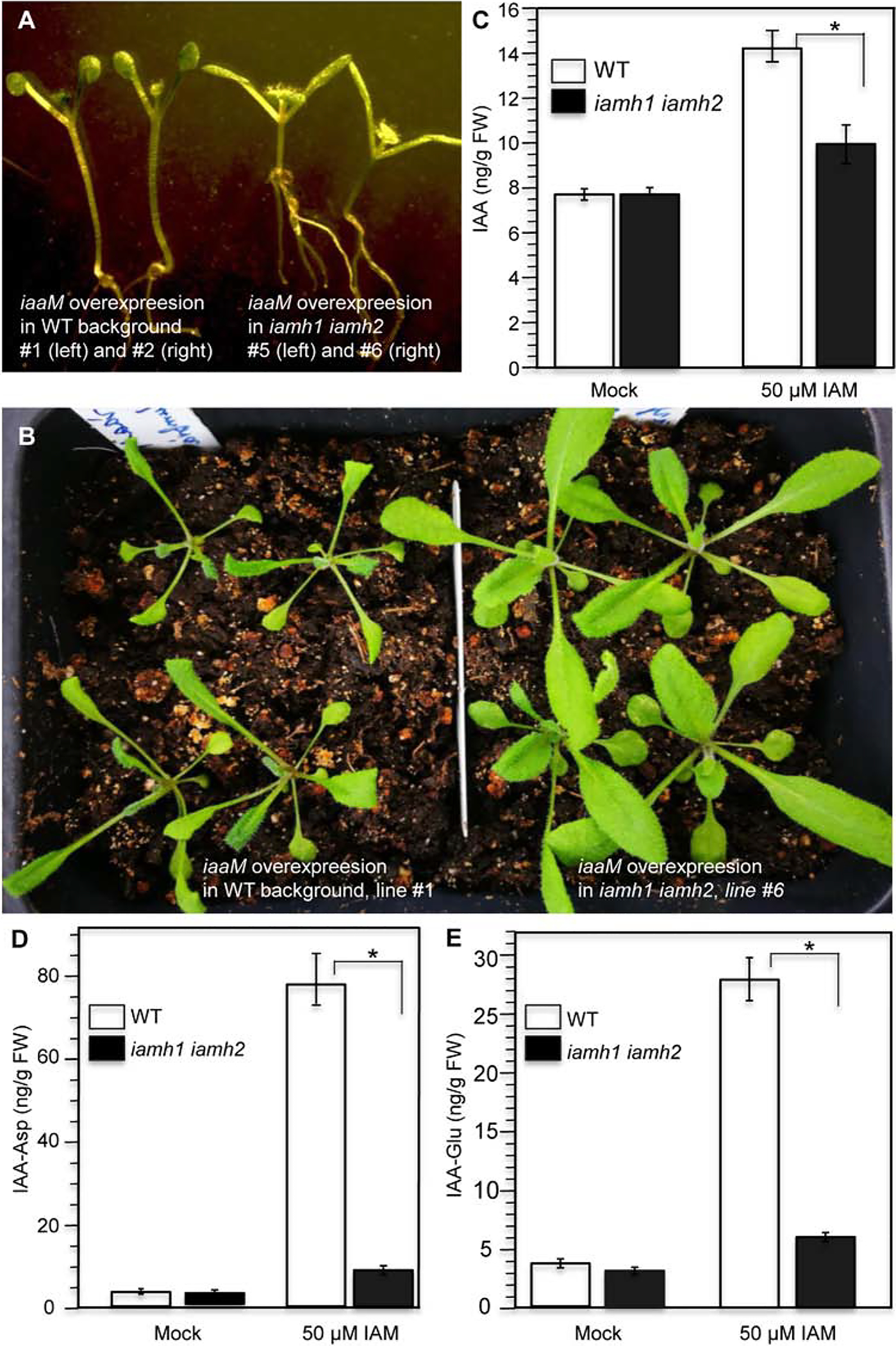
The IAMH genes are major contributors to hydrolysis of IAM in Arabidopsis. A: Disruption of the IAMH genes suppressed the iaaM overexpression phenotypes. Note that overexpression of the bacterial iaaM gene led to long hypocotyls and epinastic cotyledons (left, two independent T1 plants), which are characteristic auxin overproduction phenotypes. B: Young adult independent T1 plants of iaaM overexpression lines in WT (left) and in the iamh1 iamh2 −3 background (right). C: WT and the double mutants contained similar levels of IAA (t-value = −0.31235; P-value = 0.385191.). However, IAM treatment caused a dramatic increase of IAA in WT plants, but much less increase in the iamh1–1 iamh2–3 double mutants background (t-value = 6.55; P-value = 0.001404). D: IAM treatment caused about 80-fold increase of IAA-Asp conjugate in wild type, but only a slight increase in the iamh1–1 iamh2–3 double mutants (t-value = 19.95751; P-value = 0.000019). E: IAM treatment led to an increase in IAA-Glu synthesis in WT, but the increase was very small in the iamh1–1 iamh2–3 double mutants (t-value = 13.28318; P-value = 0.000093). * indicates statistically significant.
