-
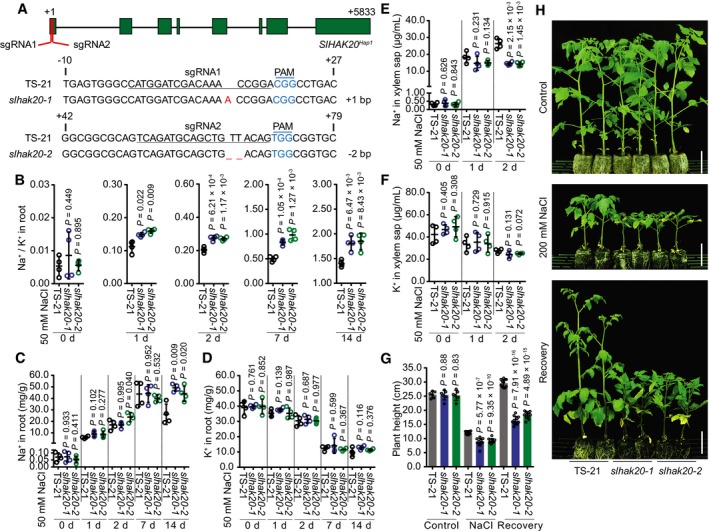
A
Generation of mutations in SlHAK20
Hap1 by CRISPR/Cas9 using two independent single‐guide RNA (sgRNA1 and sgRNA2). Sequences of SlHAK20
Hap1 in wild‐type (TS‐21), and slhak20‐1 and slhak20‐2 mutant tomato plants are shown. The sgRNA‐targeted sequences are underlined, and the protospacer adjacent motif (PAM) sequences are in blue. The deletion is indicated by a dashed line, and the insertion is labeled with red.
-
B–D
Na+/K+ ratio (B) and Na+ (C) and K+ (D) contents in roots of TS‐21 wild type and slhak20 mutants. Data are shown as means ± SD (n = 4).
-
E, F
Na+ (E) and K+ (F) contents in the xylem sap of TS‐21 and slhak20 mutants after 50 mM NaCl treatment for the indicated days. Data represent means ± SD (n = 4).
-
G
Shoot height of TS‐21, slhak20‐1, and slhak20‐2. Plant height of 24‐day‐old TS‐21, slhak20‐1, and slhak20‐2 was measured after growing under normal conditions for 14 days, followed by salt treatment for 14 days, and recovery for 10 days. Values are means ± SD (n = 10 plants of each genotype).
-
H
Salt resistance of TS‐21 wild type and slhak20 mutants. TS‐21, slhak20‐1, and slhak20‐2 grown under normal growth conditions for 24 days, followed by a treatment with 200 mM NaCl for 14 days, and then recovered for 10 days. Control plants grown under normal growth conditions for 38 days. Scale bars, 5 cm.
‐test.