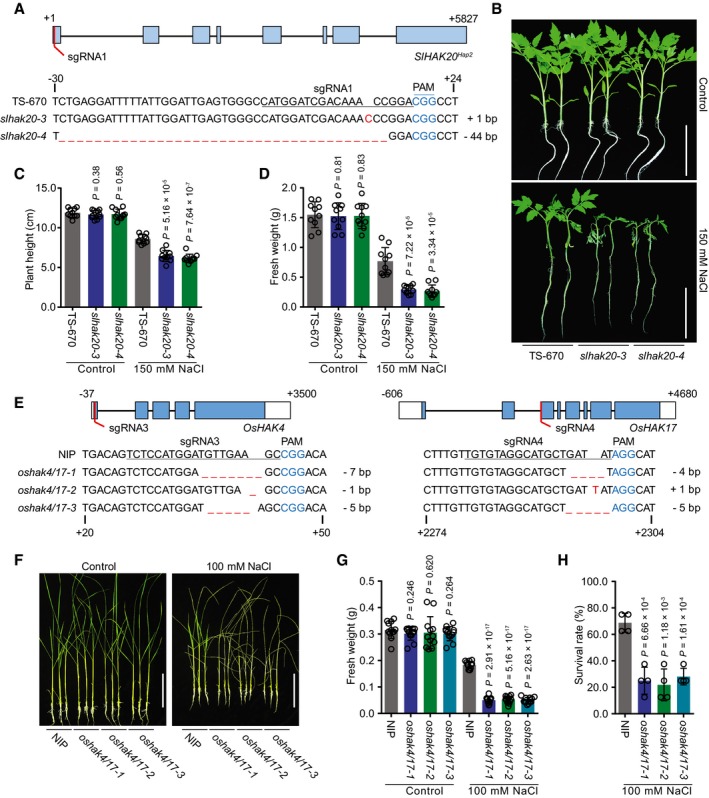
-
A
Generation of SlHAK20
Hap2 mutations using CRISPR/Cas9 system. The sequences of SlHAK20
Hap2 in wild‐type (TS‐670), and slhak20‐3 and slhak20‐4 mutant tomato plants are shown.
-
B
Salt tolerance assay of TS‐670 and slhak20 mutant plants. TS‐670, slhak20‐3, and slhak20‐4 grown under normal growth conditions for 23 days, and then treated with 150 mM NaCl for 5 days. Control plants grown under normal growth conditions for 28 days.
-
C, D
Plant height and fresh weight of TS‐670, slhak20‐3, and slhak20‐4 under normal and salt stress conditions. Shoot height (C) and fresh weight (D) of 23‐day‐old TS‐670, slhak20‐3, and slhak20‐4 were quantified after growing under normal conditions for 5 days, or salt treatment with 150 mM NaCl for 5 days. Values are means ± SD (n = 10 plants of each genotype).
-
E
Generation of oshak4/17 mutants using CRISPR/Cas9 method. Sequences of OsHAK4 and OsHAK17 in wild type (NIP), and oshak4/17‐1, oshak4/17‐2, and oshak4/17‐3 mutants are shown.
-
F–H
Phenotype of NIP and oshak4/17 double‐mutant plants under normal and salt stress conditions. 20‐day‐old seedlings of NIP and oshak4/17 mutants were treated with 100 mM NaCl for 8 days. The pictures show representative plants (F), and the fresh weight of shoots (G) and survival rate (H) were analyzed among these genotypes. Control plants were grown under normal growth conditions for 28 days, and the fresh weights of shoots were measured. Values are means ± SD (n = 12 plants of each genotype for fresh weight, four replicates for survival rate).
Data information: In (A) and (E), sgRNA‐targeted sequence and a protospacer adjacent motif (PAM) are highlighted with underline and in blue, respectively. Deletion and insertion are represented by dashed line and in red, respectively. In (C, D) and (G, H),
P‐values were determined by Student's
t‐test. Scale bar in (B) and (F), 10 cm.