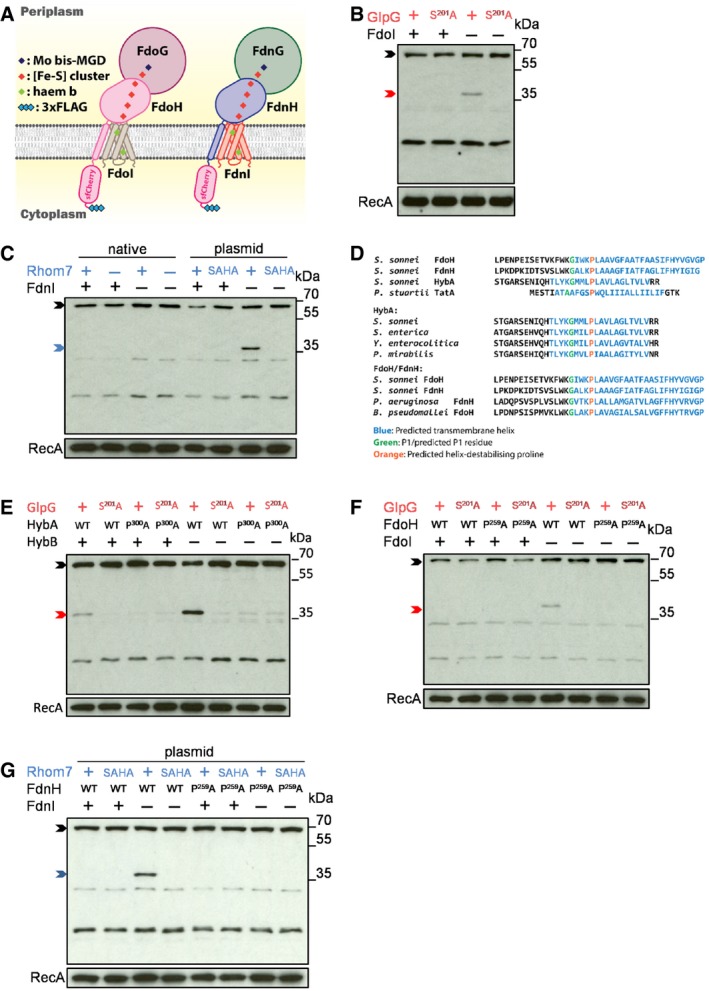
Schematic of FdoH and FdnH fusions.
Western blot analysis (probing with an anti‐FLAG mAb) to detect cleavage of FdoH cleavage in S. sonnei Δrhom7 with (+)/without (−) FdoI.
Western blot analysis to detect cleavage of FdnH in S. sonnei ΔglpG with (+) or without (−) chromosomal Rhom7 (native) or wild‐type (+)/inactive (SAHA) Rhom7 expressed from pBAD33 in S. sonnei ΔglpGΔrhom7 with (+) or without (−) FdnI.
Alignments of HybA, FdoH and FdnH in S. sonnei with Providencia stuartii TatA and with their homologues from phylogenetically diverse organisms (S. sonnei, Salmonella enterica, Yersinia enterocolitica, Proteus mirabilis, Pseudomonas aeruginosa and Burkholderia pseudomallei) highlighting conserved glycine (green) and proline (orange) residues.
Western blot analysis to detect cleavage of wild‐type (WT) or uncleavable (P300A) HybA by active (+) or inactive (S201A) chromosomal GlpG in the presence (+) or absence (−) of HybB.
Western blot analysis to detect cleavage of wild‐type (WT) or modified (P259A) FdoH by active (+) or inactive (S201A) chromosomal GlpG in the presence (+) or absence (−) of FdoI.
Western blot analysis to detect cleavage of wild‐type (WT) or modified (P259A) FdnH with active (+) or inactive (SAHA) Rhom7 expressed from pBAD33 with (+) or without (−) FdnI.
Data information: In (B, C, E‐G), rhomboid substrates that are uncleaved, cleaved by GlpG or cleaved by Rhom7 are marked by black, red and blue arrows, respectively. RecA, loading control.
Source data are available online for this figure.