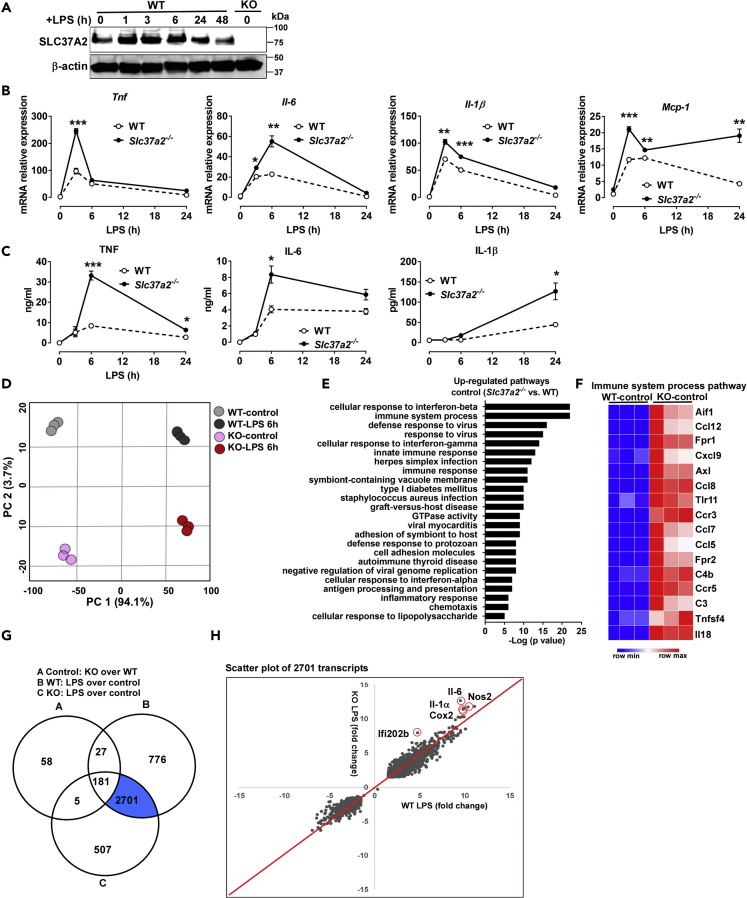
Figure 1.
Deletion of SLC37A2 Promotes Macrophage Pro-Inflammatory Activation
(A) SLC37A2 protein expression in WT bone marrow-derived macrophages (BMDMs) treated with 100 ng/mL LPS for 0–48 h, measured by western blotting. BMDMs from Slc37a2−/− mice were used as a negative control.
(B and C) Cytokine mRNA and protein expression in WT and Slc37a2−/− BMDMs stimulated with 100 ng/mL LPS for 0–24 h, measured by qPCR and ELISA, respectively.
(D) Principal component analysis of the normalized RNA-seq abundance data (transcripts per million) from BMDMs treated with or without 100 ng/mL LPS for 6 h.
(E) The top up-regulated pathways in Slc37a2−/− versus WT BMDMs in the absence of LPS stimulation, generated from pathway enrichment analysis.
(F) Heatmap of genes in the top most enriched pathway “immune system process pathway” shown in (E).
(G) A three-way Venn diagram displaying the numbers of unique and shared transcripts with log2 fold change of expression over 1.5 in Slc37a2−/− versus WT macrophages at the baseline, WT macrophages treated with LPS versus control, and Slc37a2−/− macrophages treated with LPS versus control.
(H) Scatterplot of LPS-induced transcripts. Each dot represents a gene. The x and y axis represent the LPS-induced gene log2 fold change in WT and Slc37a2−/− macrophages, respectively.
KO, Slc37a2−/−. Data are representative of three independent experiments with three samples per group (mean ± SEM). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, unpaired, two-tailed Student's t test.