Figure 3.
SLC37A2 Deletion Broadly Rewires Macrophage Metabolism at Both Transcriptional and Metabolic Levels
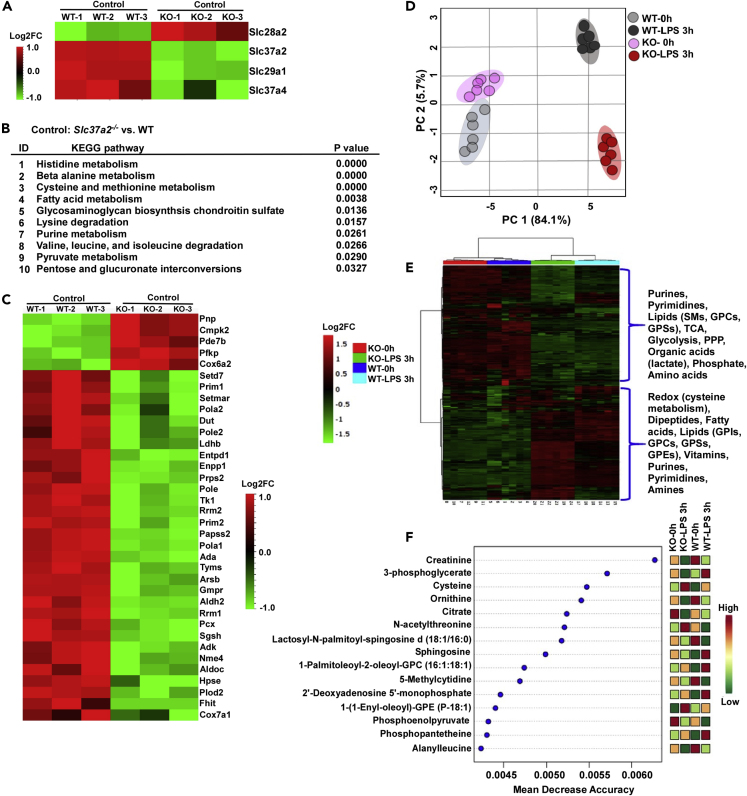
(A) Heatmap of genes in the pathway of carbohydrate derivative transporters from resting WT and Slc37a2−/− BMDMs, assessed by RNA-seq (three biologically independent samples per group).
(B) The enriched metabolic KEGG pathways in resting Slc37a2−/− versus WT BMDMs are shown and ranked according to p values, assessed by RNA-seq.
(C) Heatmap of genes under the category of metabolism in resting WT and versus Slc37a2−/− BMDMs, determined by RNA-seq.
(D) Score plots of principal component analysis of the untargeted metabolomics data collected using WT and Slc37a2−/− BMDMs treated with 100 ng/mL LPS for 0 or 3 h (n = 6 per group).
(E) Heatmap displaying hierarchical clustering analysis of metabolites in WT and Slc37a2−/− BMDMs treated with 100 ng/mL LPS for 0 or 3 h.
(F) Results of random forest analysis showing mean decrease accuracies of the top 15 metabolites altered in WT and Slc37a2−/− BMDMs treated with 100 ng/mL LPS for 0 or 3 h.
KO, Slc37a2−/−.