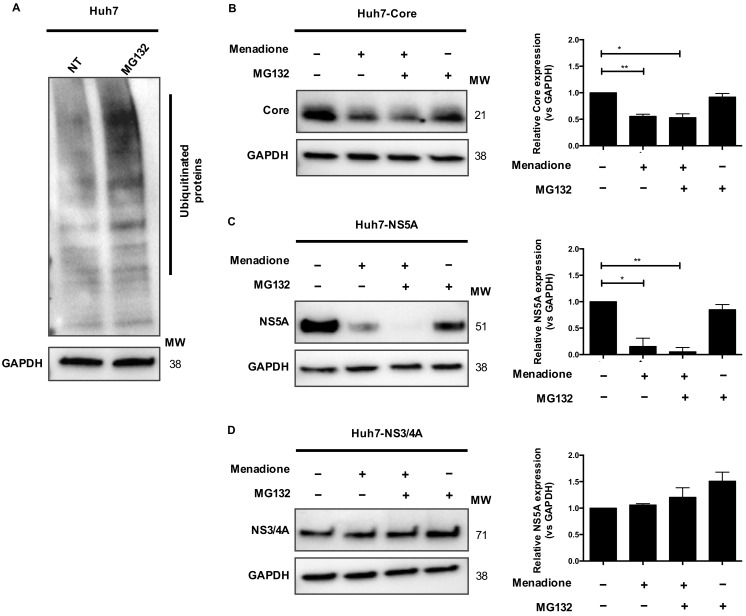
Figure 6.
Ubiquitin-proteasome pathway is not involved in HCV Core and NS5A degradation. Huh7 cells were plated in 6-well plates and treated with 10 μmol/L MG132 for 3 h. After treatment, Western blotting was performed to detect ubiquitinated proteins, GAPDH was used as loading control (A). Huh7 cells expressing HCV Core, NS3/4A, and NS5A were treated with menadione (50 μmol/L) (+/−). In some experiments 10 μmol/L MG132 was added for 3 h followed by addition of menadione (+/+). The expression of HCV Core (B), NS5A (C) and NS3/4A (D) was determined by Western blotting. GAPDH was used as a loading control. The relative protein expression was calculated based on the expression of GAPDH and compared to the expression of the control cells using ImageLab software (BioRad). The graphs depict means ± SD of three independent experiments. t test was performed to compare the means and the asterisks represent the p value ** ˂0.0039 and * ˂0.0163. (p values > 0.05 are considered not statistically significant).