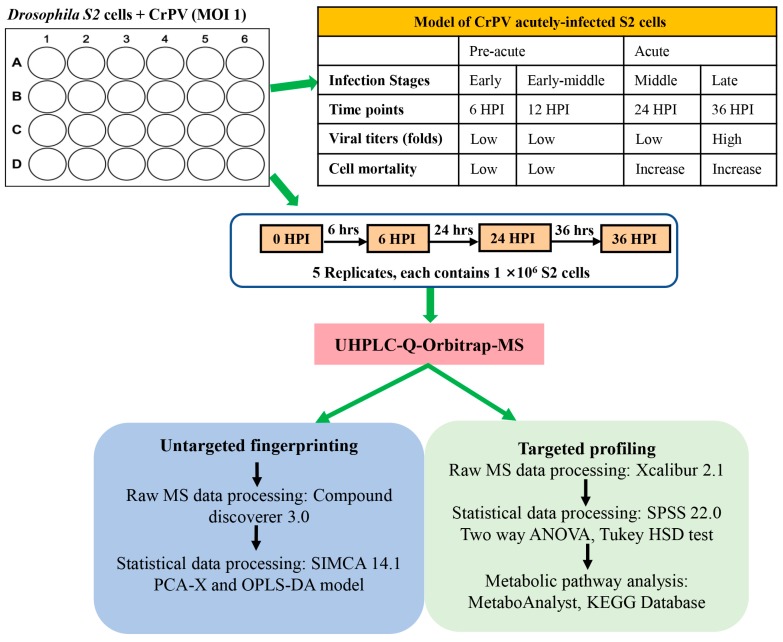
Figure 1.
Experimental design and work flow of the metabolomics study. S2 cells were infected with Cricket paralysis virus (CrPV) at MOI (multiplicity of infection) 1 and cellular extracts (5 replicates) were prepared at different times after infection. Cellular extracts were processed by ultra-high performance liquid chromatography hyphenated to quadrupole-orbitrap high-resolution mass spectrometry (UHPLC-Orbitrap-MS) for both untargeted and targeted metabolomics. In untargeted metabolomics (or fingerprinting), all possible metabolites that were differentially present in experimental samples were determined by principal component analysis (PCA-X) and orthogonal partial least square-discriminant analysis (OPLS-DA) modeling. In targeted metabolomics (or profiling), the differential presence of a collection of approximately 300 known metabolites among experimental samples was determined directly. Pathway analysis was performed on identified metabolites that significantly changed in abundance among different experimental samples.