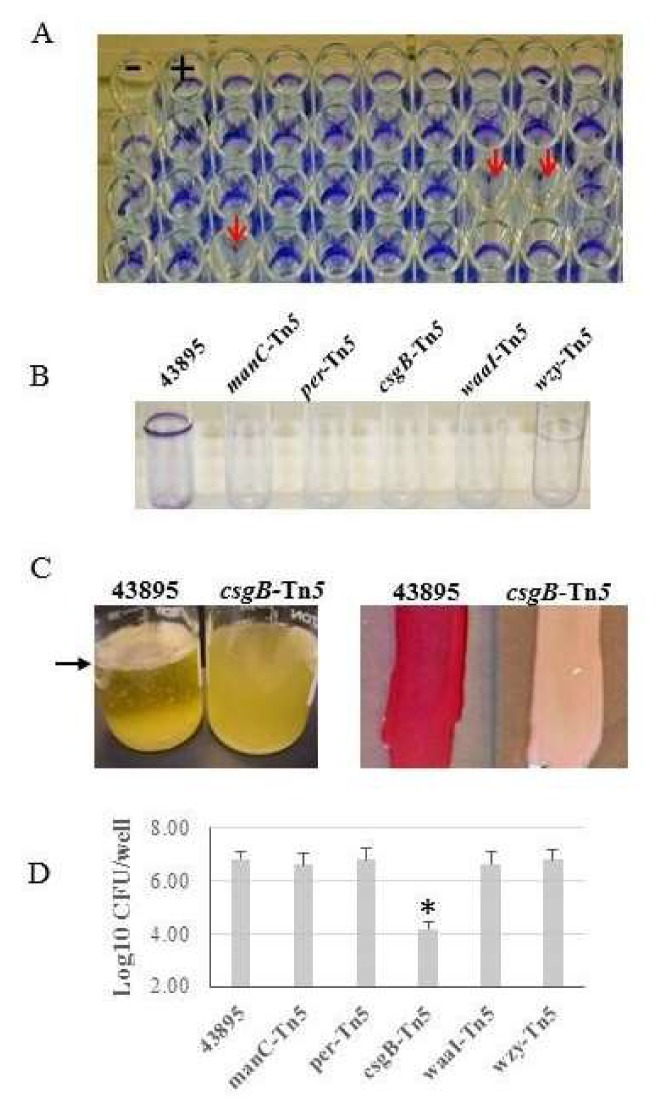
Figure 1.
O157 mutants lose biofilm, Congo red binding, and epithelial cell invasion ability. (A) A Tn5 insertion library of O157 strain 43895 (43895) was generated and screened using a crystal violet assay at 30 °C. A representative microtiter plate with Tn5 mutants is shown with control strains 43894 (biofilm-negative, -) and 43895 (parental biofilm-positive, +); arrows indicate wells with biofilm-negative mutants. (B) Biofilm-negative mutants were confirmed by a static crystal violet tube assay at 37 °C as compared to the 43895 biofilm-positive control strain. Disrupted genes were identified by sequencing across the Tn5-insertion junction as manC, per, csgB, waaI, and wzy. (C) The csgB::Tn5 mutant and the 43895 control strain were grown at 37 °C in Luria-Bertani (LB) broth without aeration to assess pellicle formation or on Congo red indicator agar to assess dye-binding. csgB::Tn5 did not form a pellicle (left) at the air-liquid interface (arrow) and had reduced Congo red dye-binding (right). (D) Epithelial cell invasion was measured by a gentamicin protection assay. Bacteria were co-cultured with bovine mammary epithelial cell line (MAC-T) monolayers (MOI = 10) and CFUs determined by plate count (triplicate experiments, three replicates/strain). Only csgB::Tn5 shows a ~100-fold reduction in cell invasion compared to 43895 and other biofilm-negative mutants; bars represent +SE and * denotes p < 0.05. The csgB::Tn5 mutant had ~10-fold reduction in adherence to MAC-T cells compared to wild type 43895 (Table 3).