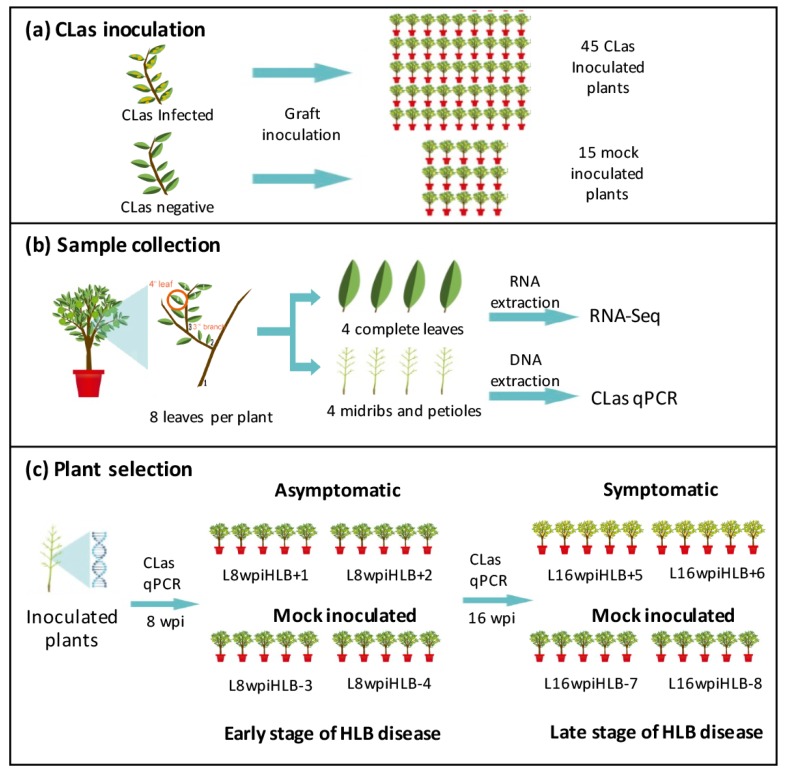
Figure 1.
Flowchart of the experimental design used to inoculate Candidatus Liberibacter asiaticus (CLas) in C. aurantifolia and to select plants to be included in the 8 libraries for RNA-seq analysis. (a) CLas inoculation by grafting. Forty-five Mexican lime plants were inoculated with CLas-positive budwood, and 15 plants were inoculated with CLas-free budwood as controls; (b) sample collection of 8 leaves per plant for RNA and DNA extraction. In all cases, the fourth leaf of 8 different branches located at third level of ramification from the main stem was collected in order to obtain a homogeneous representation of source tissue; (c) plant selection by qPCR at 8 and 16 wpi (weeks post inoculation) to construct each library. Libraries L8wpiHLB+1 and L8wpiHLB+2 included 5 pooled asymptomatic plants. Libraries L16wpiHLB+5 and L16wpiHLB+6 included 5 pooled symptomatic plants. Control libraries, L8wpiHLB-3 and L8wpiHLB-4 at 8 wpi and L16wpiHLB-7 and L16wpiHLB-8 at 16 wpi, were constructed by pooling 5 mock-inoculated plants.