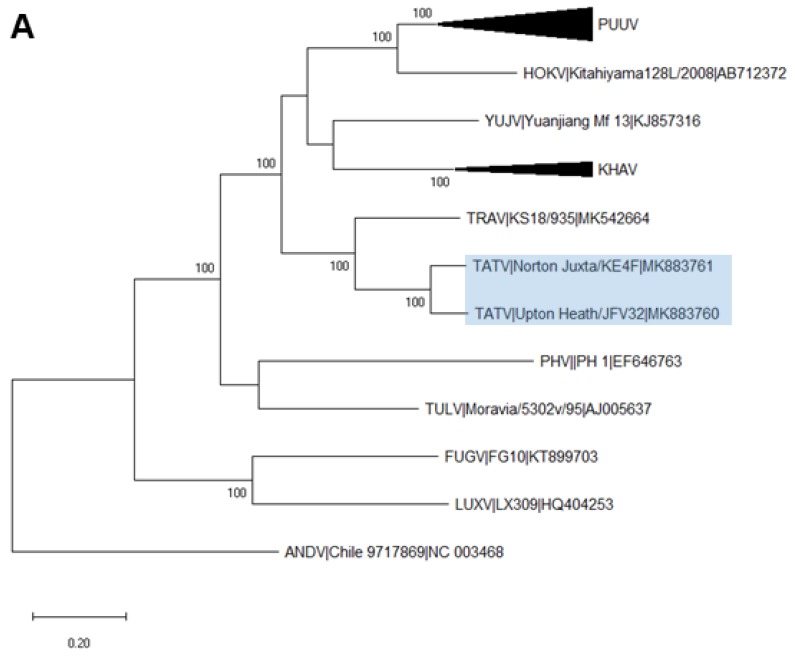
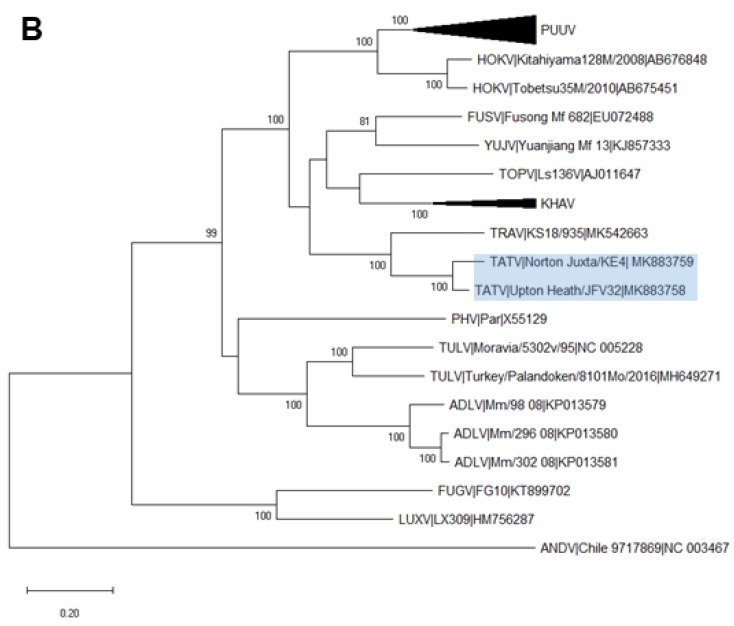
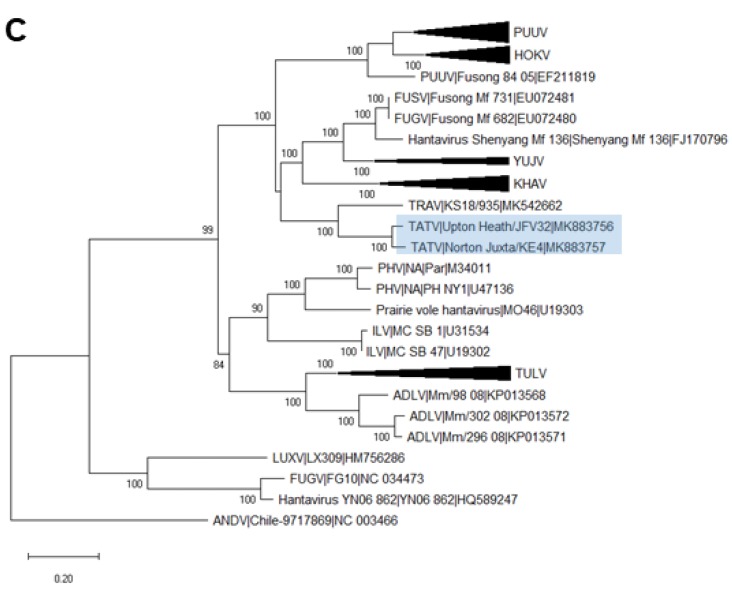
Figure 1.
Phylogenetic relationship of Tatenale virus with other vole-associated orthohantavirus species. Representative complete coding sequences were retrieved for each segment; L (A), M (B) and S (C). Maximum likelihood trees were created with a GTR+G+I model, using MEGAX software. Branch lengths were drawn to a scale of nucleotide substitutions per site. L and S trees were based on full-length sequences, whilst the M segment tree was based on the available sequence for the partial Upton-Heath strain. Numbers above individual branches show bootstrap support after 1000 replicates. Tatenale virus strains are highlighted with a blue box. Black triangles represent a compressed species-specific subtree. Sequences are shown with the species name, strain name and the GenBank accession number. PUUV, Puumala virus; HOKV, Hokkaido virus; FUSV, Fusong virus; YUJV, Yuanjiang virus; KHAV, Khabarovsk virus; TOPV, Topografov virus; TATV, Tatenale virus; TRAV, Traemmersee virus; PHV, Prospect Hill virus; ILV, Isla Vista virus; TULV, Tula virus; ADLV, Adler virus; LUXV, Luxi virus; FUGV, Fugong virus; ANDV, Andes virus.