Abstract
The stress response of 11 strains of Listeria monocytogenes to oxidative stress was studied. The strains included ST1, ST5, ST7, ST6, ST9, ST87, ST199 and ST321 and were isolated from diverse food processing environments (a meat factory, a dairy plant and a seafood company) and sample types (floor, wall, drain, boxes, food products and water machine). Isolates were exposed to two oxidizing agents: 13.8 mM cumene hydroperoxide (CHP) and 100 mM hydrogen peroxide (H2O2) at 10 °C and 37 °C. Temperature affected the oxidative stress response as cells treated at 10 °C survived better than those treated at 37 °C. H2O2 at 37 °C was the condition tested resulting in poorest L. monocytogenes survival. Strains belonging to STs of Lineage I (ST5, ST6, ST87, ST1) were more resistant to oxidative stress than those of Lineage II (ST7, ST9, ST199 and ST321), with the exception of ST7 that showed tolerance to H2O2 at 10 °C. Isolates of each ST5 and ST9 from different food industry origins showed differences in oxidative stress response. The gene expression of two relevant virulence (hly) and stress (clpC) genes was studied in representative isolates in the stressful conditions. hly and clpC were upregulated during oxidative stress at low temperature. Our results indicate that conditions prevalent in food industries may allow L. monocytogenes to develop survival strategies: these include activating molecular mechanisms based on cross protection that can promote virulence, possibly increasing the risk of virulent strains persisting in food processing plants.
Keywords: pathogen, virulence, survival, food industry, oxidizing agents, gene expression
1. Introduction
The bacterium Listeria monocytogenes is ubiquitous, and able to survive and grow at a wide range of temperatures, and in alkaline or acid media and high osmolality conditions [1]. Most of these stressful conditions are common in food processing environments (FPE) and inside the human host during infection [2]. L. monocytogenes is exposed to acid and high osmolality within food matrices (e.g. in dairy products after fermentation or in brine tanks and after addition of food preservatives) [3]. Likewise, gastric acid provides a harsh environment [4].
L. monocytogenes is exposed to diverse stresses in FPEs and during infection. Refrigeration to preserve food products both in production facilities and in consumers’ fridges imposes low temperatures, and oxidative stress is caused by sanitizer agents, especially disinfectant application and antibiotic treatments [5,6]. Disinfectants based on quaternary ammonium compounds are the most common bactericidal agents used in the food industry, and chlorine derivates or peracetic acid are also applied to prevent L. monocytogenes spread within facilities [7]. Hydrogen peroxide (H2O2) is a non-toxic, hydro-soluble and bacteriostatic or bactericidal agent also commonly used as a disinfectant [8]. Oxidizing agents cause several types of damage in cells, affecting the peptidoglycan wall and cell membrane, denaturing proteins and disrupting nucleic acid structure [6,9]. L. monocytogenes can sense stressful conditions through molecular signalling [10] and activates survival strategies to reduce oxidative damage; these strategies include expression of sigB, cold and heat shock proteins (cspABCD), proteases (clpC, clpP, groEL) and genes related to oxidative response notably superoxide dismutase (sod), perR and catalase (kat) [11,12,13]. sigB acts on genes related to stress (GRS) and virulence genes such as inlA and LIPI-1 [5]. L. monocytogenes virulence can increase under stress conditions: prfA is regulated by a sigB-depedent promoter, and clpC expression influences some genes responsible for adherence [13,14]. This relation between virulence and the stress response illustrates how L. monocytogenes may protect itself in different stressful conditions, being able to survive in environments with multiple stress factors [15].
The first aim of this study was to analyse the effect of oxidizing agents on the growth of L. monocytogenes at optimal and refrigeration temperatures. The second was to study changes in hly and clpC expression to investigate the relationship between virulence and the oxidative stress response.
2. Materials and Methods
2.1. Bacterial and Culture Conditions
Table 1 collects the information of the eleven strains used in this study that were previously isolated and characterized in Manso et al. and Melero et al. [3,16,17]. The strains of L. monocytogenes belonged to eight sequence types (ST) (ST1 (n = 1), ST5 (n = 2), ST6 (n = 1), ST7 (n = 1), ST9 (n = 3), ST87 (n = 1), ST199 (n = 1) and ST321 (n = 1)). Moreover, they were isolated from three food processing plants: six strains from a poultry meat factory, four from a dairy plant and one from a seafood company. They were found on non-food contact surfaces (n = 6), food contact surfaces (n = 2) and food (n = 3) samples. They were grown on Chromogenic Listeria Agar ISO (Oxoid, United Kingdom) at 37 °C for 48 hours. One single colony from each OCLA plates was streaked onto Tryptone Soya Agar (TSA, Oxoid) plates supplemented with 0.6% yeast extract (YE, Pronadisa, Madrid, Spain) and incubated at 37 °C for 24 hours. A single colony from each plate was used to inoculate 5 mL of Brain Heart Infusion broth (BHI broth) (Oxoid) and incubated statically overnight at 37 °C.
Table 1.
L. monocytogenes log count reduction after exposure to oxidizing agents (CHP and H2O2) at 37 °C and 10 °C.
| CHP at 37 °C a | CHP at 10 °C | H2O2 at 10 °C b | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Food Industry | Sample Type | Lineage | Strains | T1 | T2 | T3 | Stn.Error c | T1 | T2 | T3 | Stn.Error | T1 | T2 | T3 | Stn.Error |
| Mean | Mean | Mean | Mean | Mean | Mean | Mean | Mean | Mean | |||||||
| Cheese making factory | Cheese crumbs | I d | ST5 f | 5.70 | 8.35 | 9.13 * | 0.430 | 0.93 | 1.43 | 1.93 | 0.256 | 1.15 | 1.98 | 1.64 | 0.212 |
| Floor | II e | ST7 | 5.61 | 7.49 | 9.05 * | 0.611 | 1.46 | 2.01 | 2.36 | 0.592 | 0.85 | 1.54 | 2.53 | 0.752 | |
| Cheese crumbs | I | ST6 | 4.47 | 7.77 | 8.95 * | 0.148 | 1.95 | 1.60 | 1.96 | 0.597 | 1.59 | 3.47 | 4.43 | 0.691 | |
| Cheese crumbs | II | ST9 # | 6.75 | 8.99 | 9.14 * | 0.106 | 0.75 | 1.64 | 2.24 | 0.237 | 1.39 | 3.64 | 9.15 * | 0.184 | |
| Meat processing plant | Drain | I | ST87 # | 5.02 | 7.45 | 8.20 | 0.831 | 1.37 | 2.12 | 2.77 | 0.130 | 0.11 | 0.68 | 1.20 | 0.150 |
| Boxes | I | ST5 # | 5.17 | 7.57 | 9.00 | 0.251 | 1.19 | 1.67 | 2.35 | 0.098 | −0.04 | 1.85 | 2.92 | 0.627 | |
| Floor | II | ST9 # | 6.07 | 8.95 | 9.25 * | 0.171 | 1.50 | 1.96 | 2.60 | 1.150 | 3.50 | 7.15 | 9.30 * | 1.312 | |
| Wall | II | ST9 | 7.23 | 9.23 * | 9.23 * | 0.301 | 2.09 | 2.46 | 3.18 | 1.061 | 3.79 | 9.12 * | 9.12 * | 0.668 | |
| Floor | I | ST1 | 4.19 | 6.08 | 7.88 | 0.746 | 1.88 | 1.72 | 1.59 | 1.037 | 0.94 | 2.41 | 4.74 | 0.106 | |
| Drain | II | ST199 | 6.42 | 7.97 | 8.71 * | 0.621 | 2.07 | 2.68 | 4.07 | 0.495 | 3.88 | 8.62 * | 8.62 * | 0.114 | |
| Seafood company | Water machine | II | ST321 # | 7.57 | 8.92 * | 8.92 * | 0.679 | 1.57 | 3.48 | 4.43 | 0.549 | 0.97 | 3.71 | 7.97 | 0.600 |
(*) Maximum count reduction; a Cumene hydroperoxide (CHP); b Hydrogen peroxide (H2O2); c Standard error (SE); d Lineage I; e Lineage II; f Sequence type (ST). # Strains chosen to gene expression analyses by RT-PCR.
2.2. Oxidative Stress Assay
L. monocytogenes strains were grown in RPMI broth medium (1× RPMI-1640 Medium, HyClone™ and 2.05 mM L-Glutamine, GE Healthcare Life Sciences) [18] supplemented with oxidative agents according to Rea et al. [19] but with some modifications. Approximately 109 cfu/mL of the overnight culture was inoculated to 50 mL fresh medium (BHI broth) and incubated until mid-exponential phase (OD600 ~ 0.8) with shaking. After that, the overnight culture was distributed in 10 mL and centrifuged at 14,600× g for 5 min at room temperature. The bacterial pellets were collected, washed with Ringer solution (Oxoid), and centrifuged again as previously. The pellets were then resuspended in 10 mL of RPMI medium containing 8 mg/mL ferric citrate (Sigma, San Luis, Misuri, USA) and 13.8 mM cumene hydroperoxide (CHP) (Aldrich) [20], or 100 mM hydrogen peroxide (H2O2) (VWR Chemicals), or with no added agent (controls). These Listeria cultures were incubated at 10 °C and 37 °C for 4 hours, and two aliquots were taken after 2, 3 and 4 hours for enumeration and RNA extraction. Serial decimal dilutions were streaked onto TSAYE plates and were incubated at 37 °C for 24 hours to calculate the susceptibility of L. monocytogenes strains against oxidative stress conditions. All the experiments were performed in triplicate.
2.3. RNA Extraction and Gene Expression
Five out of the eleven L. monocytogenes strains (ST87, ST5 and ST9 -from a meat industry-; ST9 from a dairy plant and the ST321 from a seafood company) were chosen to perform RNA extraction using the RNA Pure Link™ RNA Mini Kit (Invitrogen, Carlsbad, California, USA) following the manufacturer’s recommendations (Table 1). RNA samples were reverse transcribed using the ImProm-II™ Reverse Transcription System (Promega, USA) as described previously [21]. Resulting cDNAs were diluted 1:20 and used as templates for specific real-time PCR assays as previously described [21] in a StepOne Real-Time PCR System (Applied Biosystems, Foster City, California, USA). Expression of hly (listeriolysin O gene) [22] and clpC (endopeptidase Clp ATP binding chain gene) [23] was studied and ldh (lactate dehydrogenase gene) [21] was used for normalization results following the 2-∆∆Ct quantification method.
2.4. Statistical Analysis
A multifactor analysis of variance was used to determine the correlation between the response to each temperature and oxidizing agents in all L. monocytogenes strains. Fisher’s least significant difference (LSD) procedure was used to determine any significant differences (p values < 0.05) amongst the means between the results for the oxidative stress at 37 °C and that at 10 °C. (Stat Graphics Centurion XVI software, Stat Graphics Centurion, Madrid, Spain).
3. Results
3.1. Response to Oxidative Stress
Table 1 shows the results of the oxidative stress in the L. monocytogenes strains tested. L. monocytogenes strains, regardless of their origin or genetic background, were significantly (p < 0.05) more tolerant to oxidizing agents (CHP and H2O2) at 10 ºC than at 37 ºC. The stress response was also significantly different (p < 0.05) between CHP and H2O2, and the mean H2O2 effect was significantly higher (p < 0.05) (Table 1).
The response to oxidative stress differed between strains at the same temperature depending on the food industry origin (Table 1). The oxidative stress response to CHP at 37 ºC differed between ST5 and ST9 strains depending on the sample types and site of isolation (Table 1) although the differences overall between strains at 37 °C were not significant (p = 1). The ST9 strain isolated from a floor in a meat processing plant was more resistant to CHP at 37 °C during the first hour (reduction of 6.07 log units); however, ST9 strains isolated from a wall in the same meat factory and from cheese crumbs showed higher count reductions (6.75 and 7.23 log unit, respectively) (Table 1). Similarly, the count reduction during the first hour for the ST5 strain isolated from the meat processing plant was lower than that for the ST5 strain isolated from the dairy plant (5.17 vs 5.70 log units) and it continued to survive after 3 h (Table 1). The ST321 strain from the seafood facility and the ST9 strain from the meat factory wall were not detectable after 2 h of incubation, whereas strains from the meat processing plant, belonging to ST1 (7.88 log unit decline), ST87 (8.20 log unit decline) and ST5 (9 log unit decline), survived for 3 h (Table 1). Similarly, the stress response to CHP at 10 °C was different within ST5 and ST9 strains; the reduction for the isolates from cheese crumbs was lower than those for the isolates from the meat processing plant: 1.93 and 2.24 log unit reduction vs. 2.35 and 2.60 and 3.18 log unit reduction after 3h of incubation, respectively (Table 1). The LSD Test indicated that the ST9 strain from the wall sample (meat processing) and the ST199 strain were significantly the most susceptible (p = 0.0129) to all the other strains at 10 °C; both were the most susceptible strains at refrigeration temperature. By contrast, ST1 and ST6 strains were the most resistant to CHP at 10 °C (Table 1).
Similar to our observations for CHP, lower temperature moderated the effect of the oxidative stress; L. monocytogenes strains were more tolerant to H2O2 at 10 °C than at 37 °C regardless the origin or genetic background of the strains. The oxidative stress response in L. monocytogenes to H2O2 at 37 °C was higher than to CHP. No colonies were found after just 1 hour of incubation in H2O2 at 37 °C, except for the ST5 strain from the dairy plant and ST87 −7.26 and 6.67 log unit declines, respectively- (Data not shown). However, after incubation at 10 °C for 3 hours, H2O2 was less toxic than CHP for the L. monocytogenes strains: count reductions were between 1.20 (ST87) and > 9.30 (ST9) log units (Table 1).
3.2. Gene Expression in Oxidative Stress Conditions
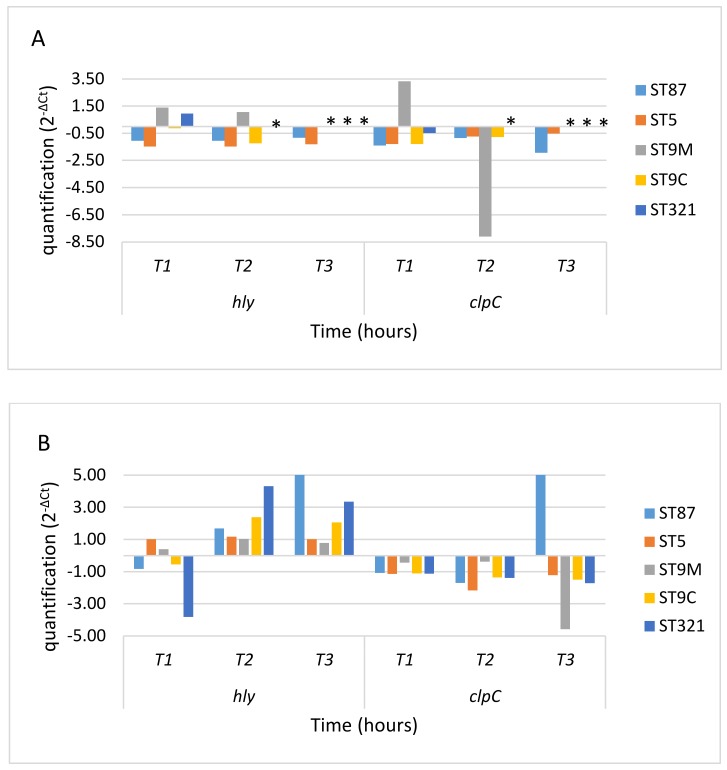
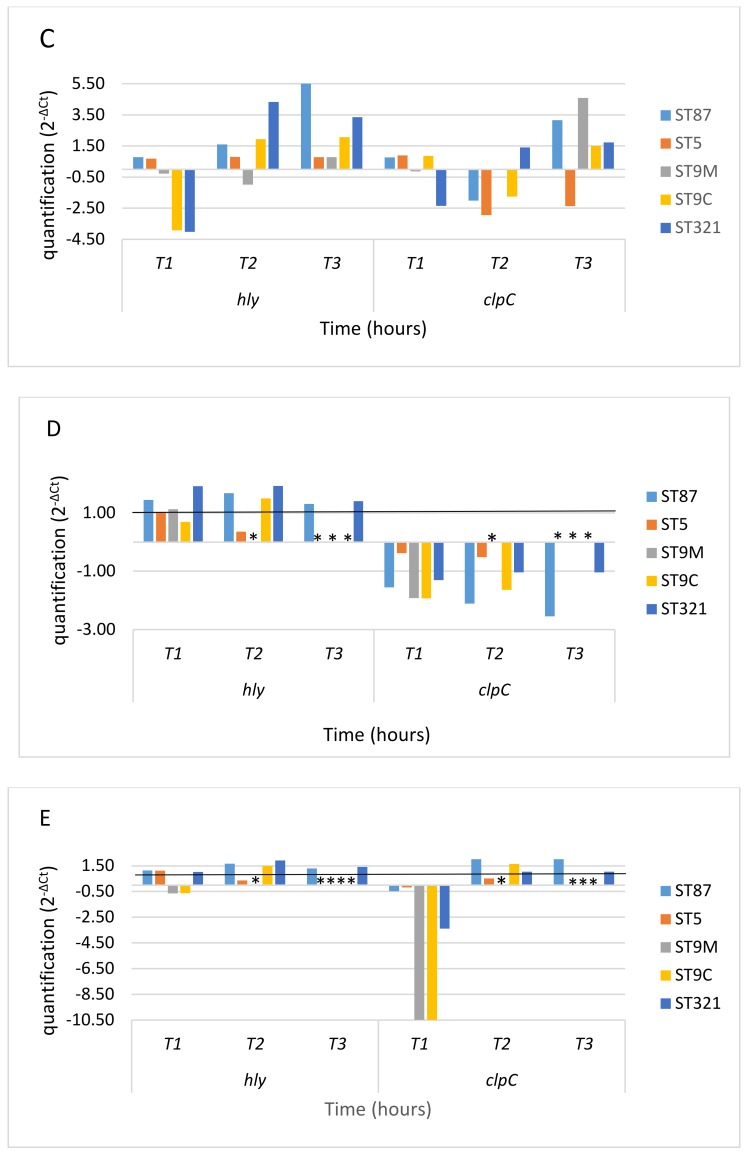
Figure 1 shows analysis of hly and clpC expression under oxidative stress conditions: hly expression was upregulated by oxidative stress (in both CHP and H2O2), and clpC was downregulated. ST9 isolated from meat and ST321 only expressed hly and clpC for the first hour in CHP at 37 °C, and there was a tendency for hly downregulation (Figure 1A).
Figure 1.
Expression of hly and clpC genes during oxidative stress. Transcripts of hly and clpC were normalized to those of the ldh gene. Expression of hly and clpC after 1, 2 and 3 hours is shown relative to that before the addition of the oxidizing agents. (A) Exposure to CHP at 37 °C; (B) Exposure to CHP at 10 °C; (C) Expression of hly and clpC in CHP at 10 °C relative to that at 37 °C; (D) Exposure to H2O2 at 10 °C; and (E) Expression of hly and clpC in H2O2 at 10 °C relative to that at 37 °C. Black line: No differences in gene expression with respect T0 (value = 1). *: gene expression results were not studied because cell counts were below the detection limit. T1, T2 and T3: period of time after 1 hour (T1), 2 hours (T2) and 3 hours (T3) of oxidative stress conditions exposure in the L. monocytogenes cultures.
Strains incubated in CHP showed higher hly expression at 10 °C than at 37 °C (Figure 1B). By contrast, clpC was downregulated in all strains tested during exposure to CHP regardless of the temperature (37 °C or 10 °C) (Figure 1A,B), with the exception of ST87 that showed clpC overexpression after CHP incubation for 3 hours at 10 °C (Figure 1B). We have also studied the relation between hly and clpC expression during CHP exposure at 10 °C and 37 °C, and it can be observed that hly expression was significantly higher when exposed to CHP at 10 °C the 37 °C, whereas clpC expression was lower under oxidative conditions regardless of the temperature (Figure 1C).
It was not possible to analyse the gene expression of hly and clpC in L. monocytogenes strains treated with H2O2 at 37 °C, because L. monocytogenes counts were below the detection limit in less than one hour. Only ST87 and ST321 survived exposure to H2O2 at 10 °C: hly was upregulated and clpC was downregulated after 3 h (Figure 1D). The relation between hly and clpC expression during H2O2 exposure at 10 °C and 37 °C was also studied (Figure 1E): only ST87 and ST321 were able to survive these oxidative conditions, but strains treated with H2O2 showed hly upregulation, while clpC was downregulated although its expression was slightly higher at 10 °C.
4. Discussion
L. monocytogenes is a foodborne bacterium commonly found in food processing plants and is able to withstand adverse conditions [1,11]. In food processing environments (FPE), various stressful conditions can influence L. monocytogenes growth and survival, especially refrigeration temperatures, osmotic, acid and oxidative stresses [24,25]. L. monocytogenes is also exposed to stressful conditions in hosts and some are common to FPE stresses: gastric acids provide acid and both invasion of phagolysosomes or macrophages and antibiotic treatments can cause oxidative stress [26,27]. L. monocytogenes is able to increase its tolerance to stressful conditions over time following repeated exposure to sub-lethal doses.
We report here that oxidative stress is temperature and oxidizing compound dependent: the effect was significantly lower at lower temperatures (10 °C vs 37 °C), and for CHP than H2O2. The role of temperature in oxidative stress has also been studied by other authors who reported that lower temperature increased the response to oxidative stress and that there was similar damage to nucleic acids and cell membranes in both stressful conditions [28,29]. In general, detergents and disinfectants (H2O2, paracetic acid and QAC compounds such as NaOCl, NH4OH4) used in food industries are applied at refrigeration temperature; this may favour the development in L. monocytogenes of resistance to these sanitizer agents over long period time [24].
Our comparison of H2O2 and CHP confirms previous reports showing that H2O2 is a more effective listericidal agent [27,30]. The presence of molecular oxygen, growth phase and serovar may all affect the response to oxidative conditions [26,31]. From the point of phenotyping strains results, we found that genotypes belonging to STs of Lineage I (ST5, ST6, ST87, ST1) were more resistant to oxidative stress than those of Lineage II (ST7, ST9, ST199 and ST321), with the exception of ST7 that showed tolerance to H2O2 at 10 °C, however the differences between strains were not statistically significant. This pattern has been observed previously: L. monocytogenes serovar 1/2a (Lineage II) is more sensitive than 4b strains (Lineage I) to 0.6 % H2O2 [24,32].
The differences between lineages may be due to difference in transcription of genes regulating and encoding oxidative responses. L. monocytogenes expresses molecular mechanisms based on stress regulator genes (sigB, ctsR, hrcA, lexA or recA) and response genes (fri, kat, perR or sod) against oxidative stress [28,32]. Most of the genes involved in stress responses in L. monocytogenes are regulated by sigB factor and they include ctsR, that is the clp operon repressor during optimal conditions. Clp family proteins (chaperones and proteases) are generally influenced by temperature [33] and stressful conditions. Likewise, clpC is also implicated in the responses to oxidative or high osmolality stresses and iron starvation [13,34]. However, kat is frequently considered the most relevant gene oxidative stress response together with sod, even at low temperature as Azizoglu & Kathariou (2010) [35] described how kat mutant strains showed smaller colonies size and less tolerance to refrigeration or freeze temperatures. In addition, the response to H2O2 by kat could be interfered by the enzymatic reaction from food products [36]. Wherefore, the present study was focused on the expression of clpC as representative gene in response to oxidative stress combined with different temperature incubation. It is well known that L. monocytogenes virulence is found in island LIPI-1, regulated by prfA [37]. Listeriolyin O is encoded by hly and its expression could be modified during exposure to range of temperature, osmotic and oxidative environmental conditions [38,39]. However, some studies supported the connection between stress conditions with virulence due to the relation among sigB and prfA, as prfA has three significant promotors dependent of sigA and sigB [14]. This inter-genetic relation could explain why stressed L. monocytogenes strains could increase their virulence, although the reaction against the stress could be different depending on possible prfA promotor sequence [40].
This study reported that clpC was overexpressed in some L. monocytogenes strains at 37 °C in the presence of both of oxidizing agents and its expression was downregulated in H2O2 at 10 °C; these findings implicate clpC in the responses to oxidative and heat stresses. Similar results were described by Ochiai et al. [28]. The relationship between stress exposure and virulence in L. monocytogenes has been studied previously. Van der Veen and Abee. [13] reported that clpC mutant strains (∆clpC) can survive inside of macrophages and other host cells; Chastanet et al. [33] found that clpP mutants were unable to grow intracellularly; and the promotors of prfA (pPrfA1 and pPrfA2) and sigB (sigA and sigB) are intrinsically regulated [34].
In conclusion, this study describes for the first time the effect of two different oxidizing agents at two temperatures (optimal growth temperature and the refrigeration temperature in food industries) at the same time on different genotypes of L. monocytogenes. The oxidative effect is temperature dependent, being lower at 10 °C than 37 °C. The virulence LIPI-1 genes were more strongly expressed when oxidative agents were applied at refrigeration temperatures.
Author Contributions
Conceptualization, D.R.-L., J.R., M.W. and B.M. (Beatriz Melero); methodology, B.M. (Beatriz Manso), B.S.;; formal analysis, D.R.-L., J.R., I.J. and B.M. (Beatriz Melero); writing—original draft preparation, B.M. (Beatriz Manso); writing—review and editing, B.M. (Beatriz Melero) and D.R.-L.; supervision, B.M. (Beatriz Melero) and D.R.-L.; supervision; project administration, I.J.; funding acquisition, D.R.-L., J.R., and M.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by European Union (EU), grant number 265877 (European 7th Framework Program).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ferreira V., Wiedmann M., Teixeira P., Stasiewicz M.J. Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. J. Food Prot. 2014;77:150–170. doi: 10.4315/0362-028X.JFP-13-150. [DOI] [PubMed] [Google Scholar]
- 2.Bergholz T.M., Shah M.K., Burall L.S., Rakic-Martinez M., Datta A.R. Genomic and phenotypic diversity of Listeria monocytogenes clonal complexes associated with human listeriosis. Appl. Microbiol. Biotechnol. 2018;102:3475–3485. doi: 10.1007/s00253-018-8852-5. [DOI] [PubMed] [Google Scholar]
- 3.Melero B., Stessl B., Manso B., Wagner M., Esteban-Carbonero Ó.J., Hernández M., Rovira J., Rodriguez-Lázaro D. Listeria monocytogenes colonization in a newly established dairy processing facility. Int. J. Food Microbiol. 2019;289:64–71. doi: 10.1016/j.ijfoodmicro.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Braschi G., Serrazanetti D.I., Siroli L., Patrignani F., De Angelis M., Lanciotti R. Gene expression responses of Listeria monocytogenes Scott A exposed to sub-lethal concentrations of natural antimicrobials. Int. J. Food Microbiol. 2018;286:170–178. doi: 10.1016/j.ijfoodmicro.2018.07.026. [DOI] [PubMed] [Google Scholar]
- 5.Pereira S.A., Alves Â., Ferreira V., Teixeira P.C.M. Listeria Monocytogenes. IntechOpen; London, UK: 2018. The impact of environmental stresses in the virulence traits of Listeria monocytogenes relevant to food safety. [Google Scholar]
- 6.Tezel U., Pavlostathis S.G. Quaternary ammonium disinfectants: Microbial adaptation, degradation and ecology. Curr. Opin. Biotechnol. 2015;33:296–304. doi: 10.1016/j.copbio.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 7.Soumet C., Ragimbeau C., Maris P. Screening of benzalkonium chloride resistance in Listeria monocytogenes strains isolated during cold smoked fish production. Lett. Appl. Microbiol. 2005;41:291–296. doi: 10.1111/j.1472-765X.2005.01763.x. [DOI] [PubMed] [Google Scholar]
- 8.Yun H.S., Kim Y., Oh S., Jeon W.M., Frank J.F., Kim S.H. Susceptibility of Listeria monocytogenes biofilms and planktonic cultures to hydrogen peroxide in food processing environments. Biosci. Biotechnol. Biochem. 2012;76:2008–2013. doi: 10.1271/bbb.120238. [DOI] [PubMed] [Google Scholar]
- 9.Harter E., Wagner E.M., Zaiser A., Halecker S., Wagner M., Rychli K. The novel stress survival islet 2 (SSI-2), predominantly present in Listeria monocytogenes strains of ST121, is involved in alkaline and oxidative stress response. Appl. Environ. Microbiol. 2017;83:e00827-17. doi: 10.1128/AEM.00827-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freitag N.E., Port G.C., Miner M.D. Listeria monocytogenes from saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 2009;7:623. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Markkula A., Lindström M., Johansson P., Björkroth J., Korkeala H. Role of four putative DEAD-box RNA helicase genes in growth of Listeria monocytogenes EGD-e under heat, pH, osmotic, ethanol, and oxidative stresses. Appl. Environ. Microbiol. 2012;78:6875–6882. doi: 10.1128/AEM.01526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suo Y., Huang Y., Liu Y., Shi C., Shi X. The expression of superoxide dismutase (SOD) and a putative ABC transporter permease is inversely correlated during biofilm formation in Listeria monocytogenes 4b G. PLoS ONE. 2012;7:e48467. doi: 10.1371/journal.pone.0048467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van der Veen S., Abee T. Contribution of Listeria monocytogenes RecA to acid and bile survival and invasion of human intestinal Caco-2 cells. Int. J. Med. Microbiol. 2011;301:334–340. doi: 10.1016/j.ijmm.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Kuhn M., Goebel W. Molecular virulence determinants of Listeria monocytogenes. Food Sci. Technol. 2017;161:111. [Google Scholar]
- 15.Komora N., Bruschi C., Magalhães R., Ferreira V., Teixeira P. Survival of Listeria monocytogenes with different antibiotic resistance patterns to food-associated stresses. Int. J. Food Microbiol. 2017;245:79–87. doi: 10.1016/j.ijfoodmicro.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Manso B., Melero B., Stessl B., Fernández-Natal I., Jaime I., Hernández M., Wagner M., Rovira J., Rodríguez-Lázaro D. Characterization of virulence and persistence abilities of Listeria monocytogenes strains isolated from food processing premises. J. Food Prot. 2019;82:1922–1930. doi: 10.4315/0362-028X.JFP-19-109. [DOI] [PubMed] [Google Scholar]
- 17.Melero B., Manso B., Stessl B., Hernandez M., Wagner M., Rovira J., Rodriguez-Lazaro D. Distribution and persistence of Listeria monocytogenes in a heavily contaminated poultry processing facility. J. Food Prot. 2019;82:1524–1531. doi: 10.4315/0362-028X.JFP-19-087. [DOI] [PubMed] [Google Scholar]
- 18.Ciolacu L., Nicolau A.I., Wagner M., Rychli K. Listeria monocytogenes isolated from food samples from a Romanian black market show distinct virulence profiles. Int. J. Food Microbiol. 2015;209:44–51. doi: 10.1016/j.ijfoodmicro.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 19.Rea R., Hill C., Gahan C.G. Listeria monocytogenes PerR mutants display a small-colony phenotype, increased sensitivity to hydrogen peroxide, and significantly reduced murine virulence. Appl. Environ. Microbiol. 2005;71:8314–8322. doi: 10.1128/AEM.71.12.8314-8322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y., Suo Y., Shi C., Szlavik J., Shi X.M., Knøchel S. Mutations in gltB and gltC reduce oxidative stress tolerance and biofilm formation in Listeria monocytogenes 4b G. Int. J. Food Microbiol. 2013;163:223–230. doi: 10.1016/j.ijfoodmicro.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 21.Bielecka M.K. Ph.D. Thesis. University of Edinburgh; Edinburgh, UK: 2011. Exploration into the Virulence Mechanisms of Listeria. [Google Scholar]
- 22.Rodríguez-Lázaro D., Pla M., Scortti M., Mozó H.J., Vázquez-Boland J.A. A novel real—Time PCR for Listeria monocytogenes that monitors analytical performance via an internal amplification control. Appl. Environ. Microbiol. 2015;71:9008–9012. doi: 10.1128/AEM.71.12.9008-9012.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H., Marquis H., Boor K.J. Sigma B contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology. 2015;151:3215–3230. doi: 10.1099/mic.0.28070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abeysundara P.D.A., Nannapaneni R., Soni K.A., Sharma C.S., Mahmoud B. Induction and stability of oxidative stress adaptation in Listeria monocytogenes EGD (Bug600) and F1057 in sublethal concentrations of H2O2 and NaOH. Int. J. Food Microbiol. 2016;238:288–294. doi: 10.1016/j.ijfoodmicro.2016.09.027. [DOI] [PubMed] [Google Scholar]
- 25.Al-Nabulsi A.A., Osaili T.M., Shaker R.R., Olaimat A.N., Jaradat Z.W., Elabedeen N.A.Z., Holley R.A. Effects of osmotic pressure, acid, or cold stresses on antibiotic susceptibility of Listeria monocytogenes. Food Microbiol. 2015;46:154–160. doi: 10.1016/j.fm.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 26.Boura M., Keating C., Royet K., Paudyal R., O’Donoghue B., O’Byrne C.P., Karatzas K.A. Loss of sigB in Listeria monocytogenes strains EGD-e and 10403S confers hyperresistance to hydrogen peroxide in stationary phase under aerobic conditions. Appl. Environ. Microbiol. 2016;82:4584–4591. doi: 10.1128/AEM.00709-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dons L.E., Mosa A., Rottenberg M.E., Rosenkrantz J.T., Kristensson K., Olsen J.E. Role of the Listeria monocytogenes 2-Cys peroxiredoxin homologue in protection against oxidative and nitrosative stress and in virulence. Pathog. Dis. 2014;70:70–74. doi: 10.1111/2049-632X.12081. [DOI] [PubMed] [Google Scholar]
- 28.Ochiai Y., Yamada F., Yoshikawa Y., Mochizuki M., Takano T., Hondo R., Ueda F. Sequential transition of the injury phenotype, temperature-dependent survival and transcriptional response in Listeria monocytogenes following lethal H2O2 exposure. Int. J. Food Microbiol. 2017;259:52–58. doi: 10.1016/j.ijfoodmicro.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Tasara T., Stephan R. Cold stress tolerance of Listeria monocytogenes: A review of molecular adaptive mechanisms and food safety implications. J. Food Prot. 2016;69:1473–1484. doi: 10.4315/0362-028X-69.6.1473. [DOI] [PubMed] [Google Scholar]
- 30.Ferreira A., O’Byrne C.P., Boor K.J. Role of sigma B in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 2001;67:4454–4457. doi: 10.1128/AEM.67.10.4454-4457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivy R.A., Wiedmann M., Boor K.J. Listeria monocytogenes grown at 7 °C shows reduced acid survival and an altered transcriptional response to acid shock compared to L. monocytogenes grown at 37 °C. Appl. Environ. Microbiol. 2012;8:3824–3836. doi: 10.1128/AEM.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y., Morvay A.A., Shi X., Suo Y., Shi C., Knøchel S. Comparison of oxidative stress response and biofilm formation of Listeria monocytogenes serotypes 4b and 1/2a. Food Control. 2018;85:416–422. doi: 10.1016/j.foodcont.2017.10.007. [DOI] [Google Scholar]
- 33.Chastanet A., Derre I., Nair S., Msadek T. clpB, a novel member of the Listeria monocytogenes CtsR regulon, is involved in virulence but not in general stress tolerance. J. Bacteriol. 2004;186:1165–1174. doi: 10.1128/JB.186.4.1165-1174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaturongakul S., Raengpradub S., Wiedmann M., Boor K.J. Modulation of stress and virulence in Listeria monocytogenes. Trends Microbiol. 2018;16:388–396. doi: 10.1016/j.tim.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Azizoglu R.O., Kathariou S. Temperature-dependent requirement for catalase in aerobic growth of Listeria monocytogenes F2365. Appl Environ Microbiol. 2010;76:6998–7003. doi: 10.1128/AEM.01223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Møretrø T., Fanebust H., Fagerlund A., Langsrud S. Whole room disinfection with hydrogen peroxide mist to control Listeria monocytogenes in food industry related environments. Int. J. Food Microbiol. 2019;292:118–125. doi: 10.1016/j.ijfoodmicro.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 37.Vázquez-Boland J.A., Kuhn M., Berche P., Chakraborty T., Domínguez-Bernal G., Goebel W., González-Zorn B., Wehland J., Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eshwar A.K., Guldimann C., Oevermann A., Tasara T. Cold-shock domain family proteins (Csps) are involved in regulation of virulence, cellular aggregation, and flagella based motility in Listeria monocytogenes. Front. Cell Infect. Microbiol. 2017;7:457. doi: 10.3389/fcimb.2017.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee T., Jun S.H., Choi C.W., Kim S.I., Lee J.C., Shin J.H. Salt stress affects global protein expression profiles of extracellular membrane- derived vesicles of Listeria monocytogenes. Microb. Pathog. 2017;115:272–279. doi: 10.1016/j.micpath.2017.12.071. [DOI] [PubMed] [Google Scholar]
- 40.Nadon C.A., Bowen B.M., Wiedmann M., Boor K.J. Sigma B contributes to PrfA mediated virulence in Listeria monocytogenes. Infect. Immun. 2002;70:3948–3952. doi: 10.1128/IAI.70.7.3948-3952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]