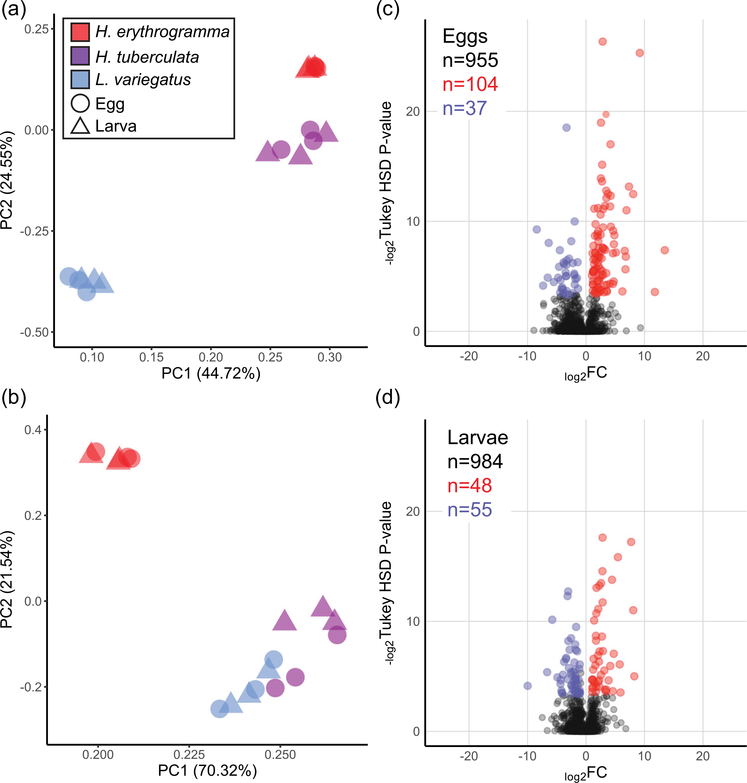
FIGURE 5.
Variation in peptide abundance is dependent on phylogeny and developmental life history. (a) When all peptides are considered (13,290 peptides encompassing 1,959 unique proteins), PC analysis of normalized spectra intensities distinguishes samples by their phylogenetic relationships (Heliocidaris vs. Lytechinus). (b) If only peptides with sequence identity across all three species are included (2,901 peptides encompassing 993 unique proteins), then most of the variation in protein content is explained by developmental life history (lecithotrophy vs. planktotrophy). This shared set of peptides was included in the differential abundance analyses between species. (c,d) Volcano plots illustrating differential relative abundance of protein molecules between (c) eggs and (d) larvae of Heliocidaris erythrogramma (red) and Heliocidaris tuberculata (blue). Only proteins that are differentially abundant between H. erythrogramma and H. tuberculata and not differentially abundant between H. tuberculata and Lytechinus variegatus are colored to mark a high confidence set of proteins potentially associated with either life history strategy. Significantly differentially abundant proteins are called as having an FC ≥ 2 and a Tukey’s honest significant difference corrected p ≤ 10%. FC: fold-change; PC: principle component