Figure 1. Maturation and Release of Peps in Response to Wounding and Pathogen Infection in Plants.
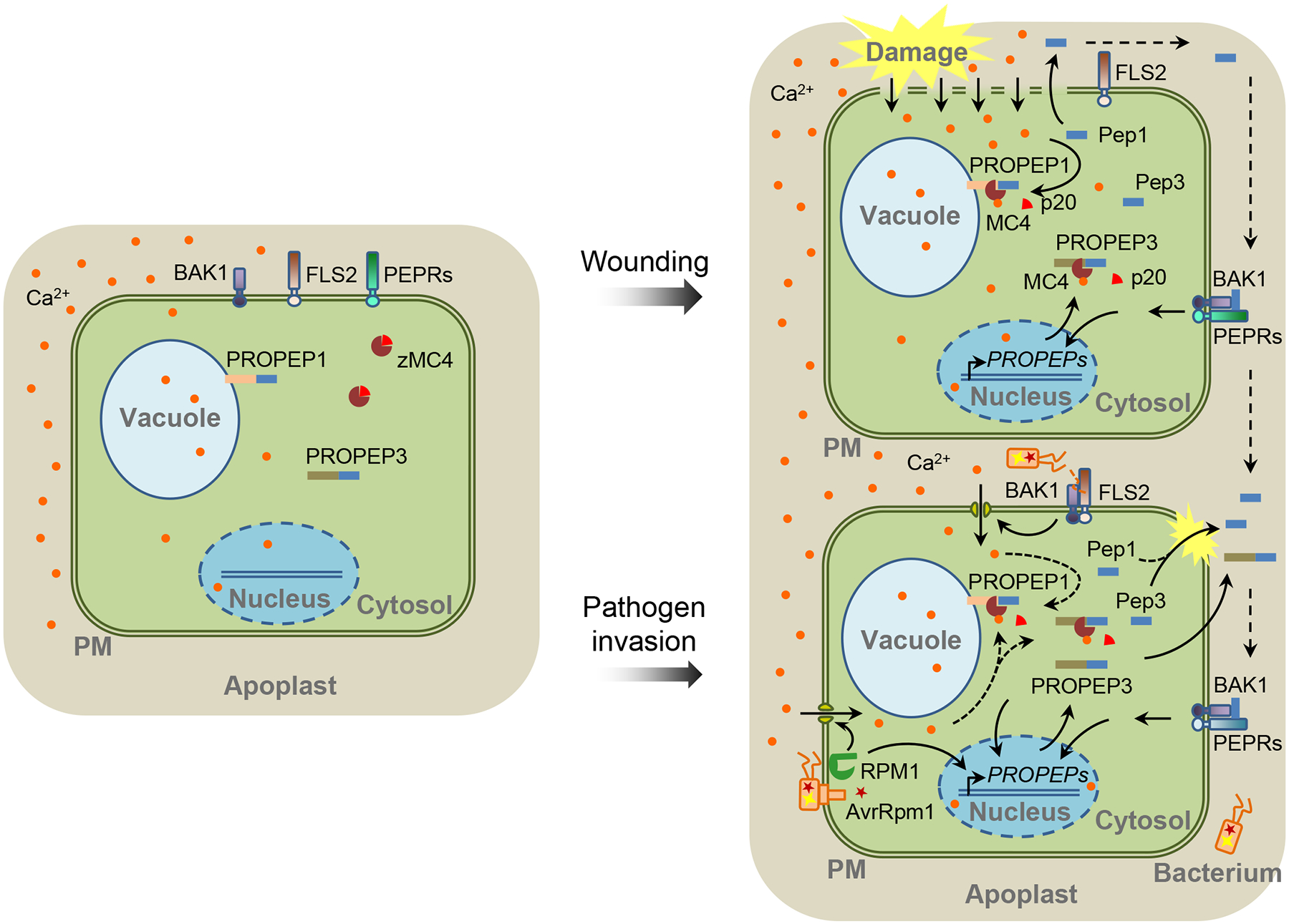
In undisturbed cells, zymogen METACASPASE 4 (zMC4), and likely other MCs, keep inactive in the cytosol. The PROPEP1 and PROPEP3 were reported to reside on the tonoplast and in the cytosol, respectively [4] (left). Cell damage leads to substantial increase of cytosolic calcium (Ca2+) ([Ca2+]cyt), which activates MC4 through autocatalytic cleavage of the p20 inhibitory domain, leading to the cleavage of PROPEPs and generation of Pep peptides. Peps as well as unprocessed PROPEPs might be released out of cells and reach the surfaces of surrounding cells. On the cell surface, they are perceived by the PEP RECEPTOR 1 (PEPR1)/PEPR2 and coreceptor BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) and induce downstream signaling events, including the transcription of PROPEP genes (right top). Upon the bacterial pathogen infections, plant cells induce an increase of [Ca2+]cyt and PROPEP transcription through the perception of pathogen-derived PATHOGEN-ASSOCIATED MOLECULAR PATTERNs (PAMPs) (such as flagellin) or effectors (such as AvrRpm1) by receptor proteins (such as the flagellin receptor FLAGELLIN SENSING 2 [FLS2] and the AvrRpm1 receptor RESISTANCE TO P. SYRINGAE PV MACULICOLA 1 [RPM1]). The [Ca2+]cyt increase may potentially activate MC4-mediated cleavage of PROPEPs (right bottom). PM: plasma membrane.
