Abstract
Despite an abundance of information in clinical genetic testing reports, information is oftentimes not well documented/utilized for decision making. Unstructured information in genetic reports can contribute to long-term patient management and future translational research. Thus, we proposed a knowledge model that could manage unstructured information in medical genetic reports and facilitate knowledge extraction, curation and updating. For this pilot study, we used a dataset including 1,565 cancer genetics reports of Mayo Clinic patients. We used a previously developed, data-driven discovery pipeline that involves both semantic annotation and co-occurrence association analysis to establish a knowledge model. We showed that compared to genetic reports, around 56% of testing results are missing or incomplete in the clinical notes. We built a genetic report knowledge model and highlighted four key semantic groups including “Genes and Gene Products” and “Treatments”. Coverage of term annotation was 99.5%. Accuracies of term annotation and relationship extraction were 98.9% and 92.9% respectively.
Introduction
Large-scale cancer genomics studies have substantially advanced our understanding of common oncology pathways and genetic alternations, and have benefited many novel therapeutic developments that target particular genetic alterations. In addition, advances in sequencing technology have also made genetic panel testing a practical option to examine genetic variants with well-known cancer treatment options1, 2. Several oncology drugs have become standards of care with companion genetics indications, e.g. trastuzumab for human epidermal growth factor receptor type 2 (HER2) breast cancer3 and vemurafenib for melanomas that have mutated BRAF4. Given the potential benefits of targeting individual patients’ tumors, i.e. individualized medicine, genetics testing panels are increasingly ordered by oncologists to facilitate decision-making during the creation of patients’ treatment plans.
Despite the abundance of information in clinical genetic testing reports, oftentimes only clinically actionable mutations validated by existing evidence are included in the summary for treatment recommendations. Other information, particularly that which is found in the unstructured text sections of genetic reports receives little attention by oncologists despite containing rich information and knowledge (disease mechanism, altered pathway, etc.) for long-term and future clinical decision support. For example, knowledge in the field of cancer genomics is accumulating at such a rapid speed that during the time between literature review and drafting of new guidelines for lung cancer treatment decisions with targeted inhibitors, major new discoveries were published for treating BRAF-mutant lung cancers and for the use of immunotherapies5. Since those guidelines are not updated frequently5-7, it is difficult for oncologists to keep up with the most current knowledge about treatment options and patient outcome expectations. Information in genetic reports is also a one-time snapshot of knowledge at the moment when the report is written. Variants of uncertain significance (VUS) might become pathogenic and actionable variants in the future. Research by Manrai et al. showed that multiple patients received misclassified variants based on the understanding at the time of testing8. Therefore, there is a need to efficiently manage information in patient’s genetic reports so that important information can be extracted, curated and periodically updated.
Taking into consideration unstructured data and the constantly updating knowledgebase of the genomics field, successful management (i.e. extraction, curation, and updating) of information in patients’ genetic reports has the potential to efficiently and deeply characterize the genetic conditions of patients, including genetic mutations and their underlying altered pathways and biological functions. This could help oncologists match patients with optimal treatment plans or clinical trials both at the moment of the test and in the future. Moreover, structuring patients’ genetic information could enable reusing clinical data for translational, such as discovery of biomarkers predictive of drug sensitivity, identification of pathways associated with response to chemotherapies9, etc. In addition, a pre-built knowledge base or knowledge graph for clinically relevant genetic information would further catalyze artificial intelligence (AI) applications in the medical field for which appropriate knowledge models are critical before any inference can be done10-13.
To achieve the above mentioned goals, we first need a knowledge model to manage the information in patient’s genetic reports14, 15. A knowledge model is a computer interpretable model or schema that organizes entities (data) and their relationships to one another within a knowledge base or database. From the database perspective, knowledge modeling is useful for abstracting and decomposing complex concepts and can address issues related to data integration and data curation15. Bimba et al.14 concluded that knowledge modeling techniques can be categorized into four groups: 1) linguistic knowledge models such as FrameNet16, WorldNet17 and ConceptNet18, which represent knowledge as lexical and semantic relationships; 2) the expert knowledge model that represents knowledge as logical and fuzzy rules19, 20; 3) ontologies that represent knowledge as taxonomies of concepts21-23; and most recently 4) the cognitive knowledge model that mimics human learning and knowledge memorization through concept algebra24. There have been several attempts to create a template for genetic reports25-27. However, all these efforts rely heavily on expertise and manual drafting. There lacks a data-driven way to construct a knowledge model for genetic reports.
In this work, we first compared the information capture rate of genetic testing results from genetic reports to clinical notes. Then we analyzed the sublanguage patterns of unstructured text sections in 1,565 oncology genetics reports. Next, we mapped extracted terms to Unified Medical Language System (UMLS) semantic types and devised a knowledge model through a data-driven discovery pipeline28 given regrouped UMLS semantic groups. Finally, we evaluated the concept coverage of the knowledge model. We believe that using our proposed data-driven method to construct a genetics knowledge model is superior to conventional and manual ways of curating domain knowledge by trained experts because it can be fully automatic, objective and scalable for a much larger genetics report corpus.
Methods
1. Comparison of Genetic Testing Results in Reports and Information Recorded in Clinical Notes
Our dataset included 1,565 cancer genetics reports (by Foundation Medicine, Inc.) from 2006 to 2018 of Mayo Clinic patients with research authorization. This research project was reviewed by the Mayo Clinic Institutional Review Board. We examined the percentage of genetic testing information recorded in clinical notes retrieved from the Mayo Clinic clinical data warehouse using a cohort of 189 gynecology (breast, ovary, cervix, and uterus) cancer patients. Based on the genetics reports, we created a list of genes from all genes identified as pathogenic or VUS. From clinical notes of the same patients, we used the list of genes to identify sentences that contained those gene names. We compared genetic test information recorded in clinical notes with original genetic test results in the reports by extracting gene name mentions from the notes. Gene name extractions were achieved using the natural language processing (NLP) system MedTagger29. MedTagger enables a series of NLP processes including regular expression matching and identification of positive/negative/possible gene name mentions with ConText30, 31. Mapping of names is insensitive to upper/lower case. MedTagger is also able to determine if the extracted keywords are related to particular patients or their family members.
2. Construction of a Knowledge Model using Patients’ Genetic Reports
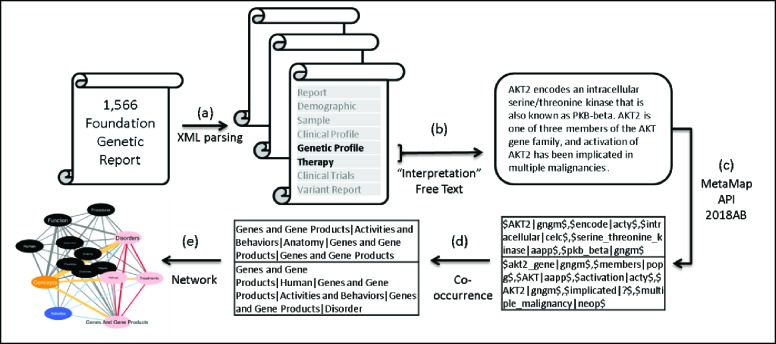
We organized text content from genetic reports according to eight originally provided major content categories: Report, Demographic, Sample (including specimen type, specimen site, ordering MD, pathologist, etc.), Clinical Profile (including submitted diagnosis and prior tests), Genetic Profile (including mutated genes, genetic variant information and VUS), Therapy, Clinical Trials, and Variant Report (Figure 1a). Additionally, there were several sub-sections in each content category. Within the “Genetic Profile” and “Therapy” categories, we first retrieved unstructured information-rich “Interpretation” sections in those reports (Figure 1b). Those sections included interpretations of biological functions of mutated genes and the effects of the alterations on gene functions, rationale for suggested therapies and supporting evidence for clinical treatments and ongoing trials at the time of reports.
Figure 1.
The workflow of our knowledge model discovery process. (a) XML parsing, (b) Extraction of unstructured text fields, (c) MetaMap semantic annotation, (d) Regrouping and co-occurrence analysis, (e) Network formation.
With rich textual data, we established a knowledge model using a previously developed, data-driven sublanguage pattern mining pipeline28 which combines NLP and semantic network analysis. Figure 1c-1e shows the workflow of our knowledge model construction process:
Term Extraction using MetaMap API: we automatically extracted medical and genetics related terms using MetaMap API 2018AB32, 33 (Figure 1c). MetaMap API is able to combine words to form phrasal entities and then assign semantic types to those phrasal entities according to 127 UMLS semantic types34. We then combined the 127 semantic types based on UMLS-provided group mappings35 and made minor changes of grouping based on our research context to reduce ambiguity in UMLS mapping. Our final grouping included 20 semantic groups: Activities, Anatomy, Genes and Gene Products, Treatments, Chemicals, Environmental Exposure, Concepts, Function, Devices, Disorders, Disease Model, Geographic Areas, Human, Living Beings, Objects, Occupations, Organizations, Physiology, Procedures, Pathway. Our customized regrouping of UMLS semantic types differs from its original grouping mainly in two aspects: 1) we separated non-therapeutic chemicals and therapeutic molecules from “Chemicals and Drugs” and created two groups – “Chemicals” and “Treatment”; 2) we created a new group called “Pathway” by extracting all phrases containing “pathway” or “signaling”.
Identification of Co-occurrence and Network Construction: co-occurrence relationships of semantic groups that occur in the same sentence were calculated (Figure 1d). A semantic network (knowledge model) was built based on the co-occurrence of semantic groups (Figure 1e). After review of the resulting knowledge model by two medical experts, we proposed to use four most significant semantic groups (Disorder, Genes and Gene Products, Pathway, Treatment) to represent a patient’s genetic profile: 1) cancer type and type of genetic mutations (from semantic group “Disorder”), 2) altered genes (from semantic group “Genes and Gene Products”), 3) altered pathways (from semantic group “Pathway”), and 4) suggested treatment (from semantic group “Treatment”).
3. Evaluations and Selected Use Case of the Knowledge Model
The knowledge model was evaluated by two medical experts based on a random sample of 200 sentences among the entire corpus of 130,238 sentences. The process was completed in two steps. First, the automatically extracted medical and genetics related terms as well as semantic annotations of the terms (Figure 1c-1d) were examined. An annotation manual was given to the experts, which included a list of standardized gene symbols and gene names from the human gene nomenclature committee (HUGO) database36 as well as normalized drug names from RxNorm37. Missing terms were extracted manually by the experts. Then, co-occurrence inferences of relationships between terms in one sentence (Figure 1e) were validated. Inter-rater agreement, coverage and accuracy were calculated.
As use cases of our knowledge model in presenting cancer genetics knowledge, we chose all of the 529 gastrointestinal cancer reports and produced a knowledge subgraph. In addition, we demonstrated a subgraph from one synthetic individual patient case with diagnosis of Colon adenocarcinoma (CRC) for better use case representation. The synthetic patient was created by randomly selecting nine “Interpretation” sections from 150 CRC patient reports. The selected nine sections were from different genes among all 143 mutated genes in 150 reports. We then examined knowledge base and literature evidence associated with any subset of terms closely connected with each other, i.e. term clusters.
Results
1. Comparison of Genetic Testing Results in Reports and Information Recorded in Clinical Notes
We examined the percentage of genetic testing information recorded in clinical notes using a subset of 189 reports of gynecology (breast, ovary, cervix, and uterus) cancer patients. Among all patients in this cohort, 57 patients (30.1%) were tested for breast, 40 patients (20.8%) were tested for uterus, 4 patients (2.1%) were tested for cervix, and 88 patients (46.6%) were tested for ovary. In our genetic reports, there were a total of 343 genes that had either pathogenic alteration (189 genes) and/or VUS (327 genes). We used the list of 343 gene names to identify sentences from clinical notes that contained those genes and variant classifications using MedTagger. Table 1 summarizes the counts for altered genes, alteration type, and VUS for the gynecology cancer cohort. The most frequently altered genes in this gynecology cancer cohort included TP53 and PIK3CA.This aligns with previous cancer genomics studies which have reported high alteration frequencies in these genes, e.g. somatic TP53 mutation occurs in 96% ovarian cancer38, and PIK3CA harbored mutations in 45% luminal A and 29% luminal B subtype breast cancer39. The most common genetic alteration was “amplification”. This matches with the genomics understanding that ovarian and triple-negative breast cancers are commonly genome instable, harboring many oncogenic amplification events40-42.
Table 1.
Distribution of Altered Genes, Alteration Type, and VUS for Gynecology Cancer Cohort
| Gene | Count | Alteration | Count | VUS | Count |
|---|---|---|---|---|---|
| TP53 | 149 | Amplification | 412 | ATM | 31 |
| PIK3CA | 51 | Indel | 368 | TP53 | 29 |
| MYC | 28 | Frameshift | 140 | BRCA2 | 29 |
| PTEN | 28 | Loss | 49 | MSH6 | 14 |
| KRAS | 27 | Splice | 41 | ALK | 13 |
By comparing report-derived genetics data versus genetic testing information recorded in clinical notes, we found that only 84 (44%) of patients’ genetics conditions were captured in clinical notes. Table 2 lists the 10 genes that are most mentioned in clinical notes. According to each gene, we categorized the patients into three groups: “Positive” means patient has pathogenic mutation in the gene, “Negative” means patient has normal gene allele, and “Possible” means that genetic test identified a VUS for this gene. Among all 189 unique genes that harbored genetics alternations reported as pathogenic, only 56 genes were captured at least once in EHR. Among 327 VUS genes reported, only 17 genes were captured at least once in clinical notes. Taking TP53 as a single gene example, there were only 26 total positive mentions in clinical notes (Table 2), representing a significant information gap compared to the 149 TP53 pathogenic events from the genetics reports (Table 1).
Table 2.
List of Most Mentioned Genes in EHR with Variant Classifications
| GeneName | Positive | Negative | Possible |
|---|---|---|---|
| BRCA1 | 20 | 46 | 3 |
| BRCA2 | 15 | 44 | 3 |
| TP53 | 26 | 6 | 1 |
| ATM | 7 | 4 | 3 |
| KRAS | 10 | 1 | 2 |
| PIK3CA | 11 | 2 | 0 |
| PTEN | 9 | 4 | 0 |
| NF1 | 7 | 1 | 1 |
| CDH1 | 4 | 3 | 1 |
| GATA3 | 5 | 2 | 1 |
2. Construction of Knowledge Model using Patients’ Genetic Report
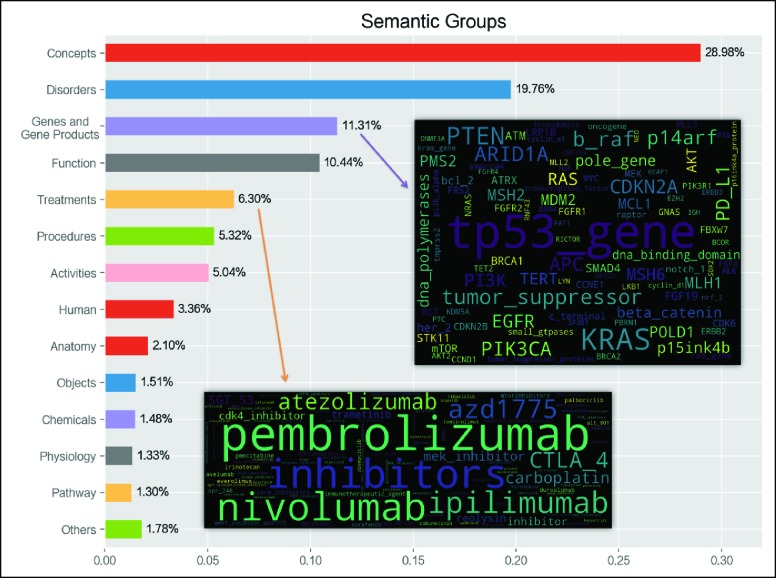
The entire corpus consisted of 130,238 sentences from 1,565 reports. 1,396,186 UMLS-identifiable terms were mapped to 115 semantic types out of 127 complete UMLS semantic types. Then, they were further regrouped to 20 semantic groups. On average, each sentence had 10.7 words. Figure 2 shows that the top seven semantic groups covered 87% of the terms in the corpus, revealing a sublanguage pattern in a confined context centered around “Disorder”, “Genes and Gene Products”, “Function”, “Treatments” and “Procedure” with “Concepts” and “Activities” being modifiers.
Figure 2.
Frequency of Terms in Major Semantic Group Reflective of Content Coverage (with word clouds for group “Genes and Gene Product” and “Treatments”)
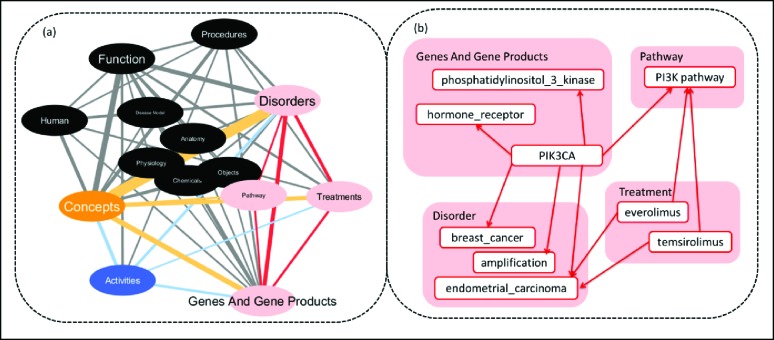
As words tend to correlate with each other over a certain range within a sentence 43, we constructed a knowledge model by calculating co-occurrence relationships between the semantic groups of terms within a sentence. As a subgraph example of the knowledge model, Figure 3a shows the top 50 most frequently co-occurred relationships represented as edges and the associated 14 semantic groups as nodes. The thickness of the edges in the network represents frequencies of co-occurrence associations between two groups. The sizes of node labels represent the number of terms in that semantic group. The graph is laid out according to edge betweenness where the betweenness of edge is defined as the number of shortest paths between node s and node t that go through e divided by the total number of shortest paths that go from s to t in the graph44, 45.
Figure 3.
(a) Knowledge model subgraph (top 50 edges, 14 nodes); Pink: key elements for a patient’s genetic profile. Orange: adjective modifies. Blue: verbal modifier. 14 semantic groups include: “Activities”, “Anatomy”, “Genes and Gene Products”, “Treatments”, “Chemicals”, “Concepts”, “Function”, “Disorders”, “Human”, “Objects”, “Physiology”, “Procedures”, “Pathway”, and “Disease Model”. (b) Expanded subgraph for term “PIK3CA”.
We selected a subset of four semantic groups to construct our knowledge model by considering semantic groups with the highest content coverage (Figure 2) and greatest relevance to clinical treatment options as measured by edge betweenness (distance between nodes shown in Figure 3a). Pink nodes are the selected nodes that represent a patient’s key genetic profile. Semantic groups “Function” and “Procedures” were not selected due to relatively large edge betweenness for edges between these two nodes and the key groups. Also terms in these two groups are less clinically relevant compared to terms in the other four groups. Orange nodes and edges represent an adjective modifier relationship. Blue nodes and edges represent a verbal modifier relationship. Figure 3b shows an example of an expanded subgraph for the term “PIK3CA”, demonstrating the power of built knowledge-base to highlight functional and pathway roles of PIK3CA. As part of PI3K signaling pathway46, PIK3CA is linked to the available targeted treatment “everolimus”47 and clinical trials related to “temsirolimus”48, 49. This subgraph is comprised of only pink nodes which are key elements of a patient’s genetic profile.
3. Evaluations and Selected Use Case of the Knowledge Model
Entity (Term) annotations were evaluated on only the four key semantic groups: “Genes and Gene Products”, “Disorders”, “Pathway”, and “Treatments”. Coverage of terms by UMLS was 99.5%. Among all the captured terms, accuracy of term annotation against HUGO and RxNorm was 98.9%. Only a few gene names, novel or complex drug names, or abbreviations for disorders could not be accurately captured or categorized by UMLS. For example, “FGF3”, “anti-PD-L1”, and “ccRCC” were gene, treatment, and disorder terms that could not be mapped by UMLS to their respective category. Inter-rater agreement was 98.8% measured by Kappa statistics.
Relationship extraction results were also evaluated based on a subset of paired relationships among the four key semantic groups. Coverage of relationship extraction was 100% i.e. our method covered all the co-occurrence pairs in a given sentence. Based on expert evaluations, semantic accuracy of relationship extraction was 92.9%. We noticed that our method had an increased error rate on complex sentences containing multiple drugs, diseases or gene mentions with co-occurred but no direct semantic relationships. For example, “mRNA_expression” and “carcinosarcoma” in “no differences were observed in FGFR2 mRNA expression in tumors as compared to normal tissue in a study including four patients with carcinosarcoma”; “ipilimumab” and “atezolizumab” in “FDA approved agents include ipilimumab, atezolizumab, avelumab, durvalumab, pembrolizumab, and nivolumab”. Inter-rater agreement was 95.6% between the two experts.
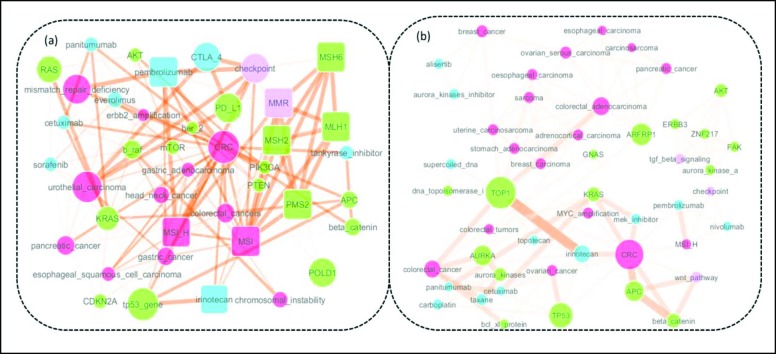
A knowledge subgraph built from 529 gastrointestinal cancer reports is shown in Figure 4a. We examined knowledge base and literature evidence associated with term clusters identified from subgraphs. For example, a mismatch repair (MMR) cluster defined as first degree neighbors connected with a central term “MMR”, was chosen from subgraph shown in the upper right area of Figure 4a. We used a square shape to represent the nodes in the cluster. In this cluster, we found that key gene members in related MMR pathway, such as “MLH1”, “MSH2”, “MSH6” and “PMS2”50, 51; in addition, we also found directly related disorder terms “MSI-H” (“microsatellite instability – high”) and “MSI” (“microsatellite instability”) caused by deficiency of MMR pathway52; in terms of treatment options, we found two treatment terms were co-clustered with “MMR”: “pembrolizumab” and “irinotecan”, which are immunotherapy and chemotherapy reported to be associated with genetic defects of MMR pathway53, 54.
Figure 4.
(a) Gastrointestinal knowledge subgraph centered on “CRC” and “colorectal cancer”. (Red: Disorder, Green: Genes and Gene Products, Blue: Treatment, Purple: Pathway) (b) Knowledge subgraph of one patient with Colon adenocarcinoma (CRC)
In addition, subgraph from one synthetic individual patient case with diagnosis of Colon adenocarcinoma (CRC) is shown as Figure 4b. We can see that this graph captured all the genetic mutations of this synthetic patient: KRAS, AURKA, MYC, GNAS, APC, ARFRP1, TOP1, TP53, and ZNF217. Closely related genomic products and pathways: “beta catenin”, “aurora kinases”, “bcl-xl protein”, “dna topoisomerase I”, “aurora kinase a”, “checkpoint pathway”, “tgf beta signaling pathway”, and “wnt pathway” were also extracted. A list of recommended therapies were also capture: “alisertib”, “aurora_kinases_inhibitor”, “cetuximab”, “carboplatin”, “mek_inhibitor”, “nivolumab”, “pembrolizumab”, “panitumumab” and “taxane”. Most importantly, the relationships between terms were also captured.
Discussions
In this pilot study, we described our investigation and initial attempt to utilize data-driven sublanguage analysis to construct a knowledge model for genetic testing report results. We identified an information capture gap between genetic reports and summarized genetic test results in clinical notes – 56% of test results were missing or incomplete in clinical notes. Previous research has shown that only information used in diagnosis or treatment decision making is captured in clinical notes55. Hence, our results indicate that although genetic tests have been increasingly used for facilitating clinical decision making processes, actual genetic results were under-represented or -captured in clinical notes. This suggests a further need to enhance capture of genetic results involved in decision making processes. The incomplete documentation might be because gene alterations identified in the tests lack supporting evidence to guide immediate treatment decisions. Even though current reports might not provide much useful information, it remains critical to continuously monitor research updates that may be relevant to an individual patient’s long-term management. Additionally, enabling data reuse for future translational research is also significant. Therefore, a comprehensive genetic knowledge model that can provide guidance for capture and structure patient genetic information from unstructured text is important.
In this study, we used MedTagger to identify gene name mentions in clinical notes and used the built-in ConText module to identify results as positive/negative/possible. However, we found the accuracy of ConText positivity identification to be low (31%), and thereby manually curated positivity for a number of results. We believe this is because ConText was built to identify the positivity of diseases or events but not test results. In some instances, gene name mentions were identified as positive when the sentences were actually referring to genes included in the testing panel or the oncologist was simply giving a general explanation of the risks of having mutations in these genes. In future work, we will update the ConText module to achieve a better performance of positivity identification.
Through our analysis of the unstructured information in genetic reports and clinical notes, we further highlighted a critical need for mapping and normalizing genes, pathways, and treatment names using a standardized nomenclature or ontology. In order to fully capture genetic variant-level information, Sequencing Ontology56 should be incorporated into the future annotation system, in addition to the UMLS, to identify mentions of biological sequence features e.g. binding site, exon.
Future work with our proposed knowledge model could involve the extraction, curation and visualization of multiple facets (gene alteration type, recommended treatment, disorder) of patient characteristics given an affected gene. We will be able to build a query portal for oncologists to identify similar patients in the database. A structured patient genetic profile created from the knowledge model could also facilitate the collection and management of relevant and up-to-date information for each patient. Curated information could also facilitate future translational research. All three potential uses could help oncologists with better decision making and long-term management given a new patient, as well as translational researchers with more structured and comprehensive dataset to work with.
Figures & Tables
References
- 1.Cardoso F, van’t Veer LJ, Bogaerts J, et al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. New England Journal of Medicine. 2016;375:717–29. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 2.Pritchard CC, Smith C, Salipante SJ, et al. ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing. The Journal of Molecular Diagnostics. 2012;14:357–66. doi: 10.1016/j.jmoldx.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. New England Journal of Medicine. 2001;344:783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 4.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. New England Journal of Medicine. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Journal of Thoracic Oncology. 2018;13:323–58. doi: 10.1016/j.jtho.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 6.El-Shanti HI, Ferguson PJ. Chronic recurrent multifocal osteomyelitis: a concise review and genetic update. Clinical Orthopaedics and Related Research®. 2007;462:11–9. doi: 10.1097/BLO.0b013e3180986d73. [DOI] [PubMed] [Google Scholar]
- 7.Riley BD, Culver JO, Skrzynia C, et al. Essential elements of genetic cancer risk assessment, counseling, and testing: updated recommendations of the National Society of Genetic Counselors. Journal of genetic counseling. 2012;21:151–61. doi: 10.1007/s10897-011-9462-x. [DOI] [PubMed] [Google Scholar]
- 8.Manrai AK, Funke BH, Rehm HL, et al. Genetic misdiagnoses and the potential for health disparities. New England Journal of Medicine. 2016;375:655–65. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mangaonkar AA, Ferrer A, e Vairo FP, et al. Clinical Applications and Utility of a Precision Medicine Approach for Patients With Unexplained Cytopenias; Mayo Clinic Proceedings; Elsevier; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi E, Xu Z, Li Y, et al. Graph Convolutional Transformer: Learning the Graphical Structure of Electronic Health Records. arXiv preprint arXiv. 2019;190604716 [Google Scholar]
- 11.Xiao H, Huang M, Zhu X. TransG: A generative model for knowledge graph embedding; Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers); 2016. pp. 2316–25. [Google Scholar]
- 12.Toutanova K, Chen D, Pantel P, Poon H, Choudhury P, Gamon M. Representing text for joint embedding of text and knowledge bases; Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing; 2015. pp. 1499–509. [Google Scholar]
- 13.Choi E, Xiao C, Stewart W, Sun J. Mime: Multilevel medical embedding of electronic health records for predictive healthcare. Advances in Neural Information Processing Systems. 2018:4547–57. [Google Scholar]
- 14.Bimba AT, Idris N, Al-Hunaiyyan A, et al. Towards knowledge modeling and manipulation technologies: A survey. International Journal of Information Management. 2016;36:857–71. [Google Scholar]
- 15.Storey VC, Song I-Y. Big data technologies and management: What conceptual modeling can do. Data & Knowledge Engineering. 2017;108:50–67. [Google Scholar]
- 16.Baker C. FrameNet: a knowledge base for natural language processing. Proceedings of Frame Semantics in NLP : A workshop in honor of Chuck Fillmore (1929-2014) 2014:1–5. [Google Scholar]
- 17.Fellbaum C. Towards a representation of idioms in WordNet. Usage of WordNet in Natural Language Processing Systems. 1998 [Google Scholar]
- 18.Speer R, Havasi C. Representing General Relational Knowledge in ConceptNet 5. LREC. 2012:3679–86. [Google Scholar]
- 19.Driankov D, Hellendoorn H, Reinfrank M. An introduction to fuzzy control. Springer Science & Business Media; 2013. [Google Scholar]
- 20.Kerr-Wilson J, Pedrycz W. Design of rule-based models through information granulation. Expert Systems with Applications. 2016. 46:274–85. [Google Scholar]
- 21.Ilyas QM, Anwar W. Contextual advertising using keyword extraction through collocation; Proceedings of the 7th international conference on frontiers of information technology; ACM; 2009. p. 69. [Google Scholar]
- 22.Sánchez D. A. methodology to learn ontological attributes from the Web. Data & Knowledge Engineering. 2010;69:573–97. [Google Scholar]
- 23.Van Heijst G, Schreiber AT, Wielinga BJ. Using explicit ontologies in KBS development. International journal of human-computer studies. 1997;46:183–292. [Google Scholar]
- 24.Wang Y. Towards the abstract system theory of system science for cognitive and intelligent systems. Complex & Intelligent Systems. 2015;1:1–22. [Google Scholar]
- 25.Scheuner MT, Hilborne L, Brown J, Lubin ftmotRMGTRAB, Ira M. A report template for molecular genetic tests designed to improve communication between the clinician and laboratory. Genetic testing and molecular biomarkers. 2012;16:761–9. doi: 10.1089/gtmb.2011.0328. [DOI] [PubMed] [Google Scholar]
- 26.Haga SB, Mills R, Pollak KI, et al. Developing patient-friendly genetic and genomic test reports: formats to promote patient engagement and understanding. Genome medicine. 2014;6:58. doi: 10.1186/s13073-014-0058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brownstein CA, Beggs AH, Homer N, et al. An international effort towards developing standards for best practices in analysis, interpretation and reporting of clinical genome sequencing results in the CLARITY Challenge. Genome biology. 2014;15:R53. doi: 10.1186/gb-2014-15-3-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y, Fesharaki NJ, Liu H, Luo J. Using data-driven sublanguage pattern mining to induce knowledge models: application in medical image reports knowledge representation. BMC medical informatics and decision making. 2018;18:61. doi: 10.1186/s12911-018-0645-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, Bielinski SJ, Sohn S, et al. An information extraction framework for cohort identification using electronic health records. AMIA Summits on Translational Science Proceedings. 2013;2013:149. [PMC free article] [PubMed] [Google Scholar]
- 30.Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. Journal of biomedical informatics. 2009;42:839–51. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapman WW, Chu D, Dowling JN. ConText: An algorithm for identifying contextual features from clinical text; Proceedings of the workshop on BioNLP 2007: biological, translational, and clinical language processing; Association for Computational Linguistics; 2007. pp. 81–8. [Google Scholar]
- 32.Aronson AR. Bethesda, MD: NLM, NIH, DHHS; 2006. Metamap: Mapping text to the umls metathesaurus; pp. 1–26. [Google Scholar]
- 33.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program; Proceedings of the AMIA Symposium; American Medical Informatics Association; 2001. p. 17. [PMC free article] [PubMed] [Google Scholar]
- 34.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32:D267–D70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCray AT, Burgun A, Bodenreider O. Aggregating UMLS semantic types for reducing conceptual complexity. Studies in health technology and informatics. 2001;84:216. [PMC free article] [PubMed] [Google Scholar]
- 36.Eyre TA, Ducluzeau F, Sneddon TP, Povey S, Bruford EA, Lush MJ. The HUGO gene nomenclature database, 2006 updates. Nucleic acids research. 2006;34:D319–D21. doi: 10.1093/nar/gkj147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S, Ma W, Moore R, Ganesan V, Nelson S. RxNorm: prescription for electronic drug information exchange. IT professional. 2005;7:17–23. [Google Scholar]
- 38.Network CGAR. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vollebergh MA, Jonkers J, Linn SC. Genomic instability in breast and ovarian cancers: translation into clinical predictive biomarkers. Cellular and molecular life sciences. 2012;69:223–45. doi: 10.1007/s00018-011-0809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watkins JA, Irshad S, Grigoriadis A, Tutt AN. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Research. 2014;16:211. doi: 10.1186/bcr3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang ZC, Birkbak NJ, Culhane AC, et al. Profiles of genomic instability in high-grade serous ovarian cancer predict treatment outcome. Clinical cancer research. 2012;18:5806–15. doi: 10.1158/1078-0432.CCR-12-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beeferman D, Berger A, Lafferty J. A. model of lexical attraction and repulsion; Proceedings of the eighth conference on European chapter of the Association for Computational Linguistics; Association for Computational Linguistics; 1997. pp. 373–80. [Google Scholar]
- 44.Yoon J, Blumer A, Lee K. An algorithm for modularity analysis of directed and weighted biological networks based on edge-betweenness centrality. Bioinformatics. 2006;22:3106–8. doi: 10.1093/bioinformatics/btl533. [DOI] [PubMed] [Google Scholar]
- 45.Girvan M, Newman ME. Community structure in social and biological networks. Proceedings of the national academy of sciences. 2002;99:7821–6. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di Nicolantonio F, Arena S, Tabernero J, et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. The Journal of clinical investigation. 2010;120:2858–66. doi: 10.1172/JCI37539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loi S, Michiels S, Baselga J, et al. PIK3CA genotype and a PIK3CA mutation-related gene signature and response to everolimus and letrozole in estrogen receptor positive breast cancer. PloS one. 2013;8:e53292. doi: 10.1371/journal.pone.0053292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janku F, Wheler JJ, Westin SN, et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. Journal of clinical oncology. 2012;30:777. doi: 10.1200/JCO.2011.36.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fleming GF, Ma CX, Huo D, et al. Phase II trial of temsirolimus in patients with metastatic breast cancer. Breast cancer research and treatment. 2012;136:355–63. doi: 10.1007/s10549-011-1910-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonadona V, Bonaïti B, Olschwang S, et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. Jama. 2011;305:2304–10. doi: 10.1001/jama.2011.743. [DOI] [PubMed] [Google Scholar]
- 51.Prolla TA, Baker SM, Harris AC, et al. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DMA mismatch repair. Nature genetics. 1998;18:276. doi: 10.1038/ng0398-276. [DOI] [PubMed] [Google Scholar]
- 52.Boyer JC, Umar A, Risinger JI, et al. Microsatellite instability, mismatch repair deficiency, and genetic defects in human cancer cell lines. Cancer research. 1995;55:6063–70. [PubMed] [Google Scholar]
- 53.Kim JE, Hong YS, Ryu MH, et al. Association between deficient mismatch repair system and efficacy to irinotecan‐containing chemotherapy in metastatic colon cancer. Cancer science. 2011;102:1706–11. doi: 10.1111/j.1349-7006.2011.02009.x. [DOI] [PubMed] [Google Scholar]
- 54.Prasad V, Kaestner V, Mailankody S. Cancer drugs approved based on biomarkers and not tumor type—FDA approval of pembrolizumab for mismatch repair-deficient solid cancers. JAMA oncology. 2018;4:157–8. doi: 10.1001/jamaoncol.2017.4182. [DOI] [PubMed] [Google Scholar]
- 55.Fu S, Leung LY, Wang Y, et al. Natural Language Processing for the Identification of Silent Brain Infarcts From Neuroimaging Reports. JMIR medical informatics. 2019;7:e12109. doi: 10.2196/12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eilbeck K, Lewis SE, Mungall CJ, et al. The Sequence Ontology: a tool for the unification of genome annotations. Genome biology. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]