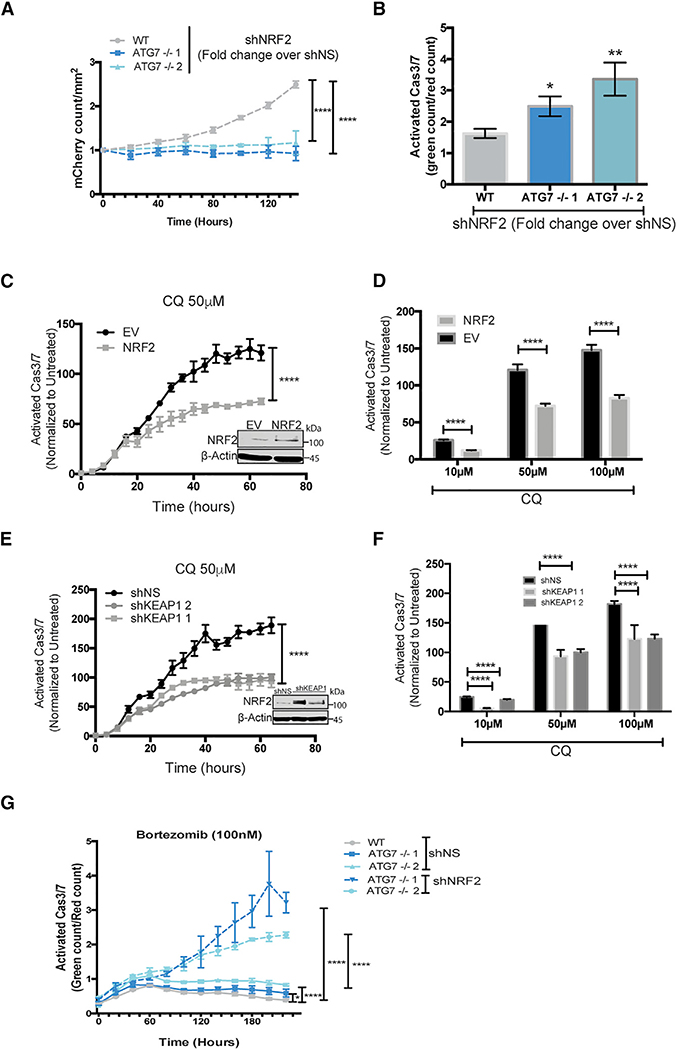
Figure 7. ATG7−/− Clones Are Dependent on NRF2 for Survival.
(A–G) Incucyte live cell imaging of (A and B) BT459 cells transduced with shNS or shNRF2. (A) KD of NRF2 induced effects on growth. Data are represented as mean ± SD of the fold change in Incucyte mCherry+ cell count/mm2 for shNRF2 over shNS for technical replicates (N = 3) representative of 2 individual experiments. Statistical analysis: two-way ANOVA.
(B) KD of NRF2 induced effects on apoptosis 8 days after transduction with indicated shRNAs measured as the fold change in CellEvent green count for shNRF2 relative to shNS. Data are represented as mean ± SD for technical replicates (N = 3) representative of 2 individual experiments. Statistical analysis: one-way ANOVA.
(C–G) Normalized Caspase3/7 Cell Event green in (C–F) parental BT549 mCherry-NLS cells with (C and D) stable overexpression of EV or NRF2-flag or (E and F) shNS or shRNAs targeting KEAP1 or (G) BT549 WT or ATG7−/− with KD of NRF2.
(C and E) Data represented as the mean fold change in treated over untreated cells ± SEM over time or (D and F) 3 days after treatment with indicated doses of CQ for technical replicates (N = 3) and representative of 2 individual experiments. Statistical analysis: two-way ANOVA. Inset: western blot of stable lines overexpressing NRF2.
(G) After transduction with shNS or shNRF2 and treatment with bortezomib (100 nM) at indicated time points. Data are represented as the mean ± SEM for technical replicates (N of 3) and the graphs shown are representative of 2 individual experiments. Statistical analysis: two-way ANOVA. *p % 0.05, **p % 0.01, ***p %0.01, ****p % 0.001, *****p % 0.0001, significance indicated for the last time point for time course graphs. See also Figures S6 and S7.