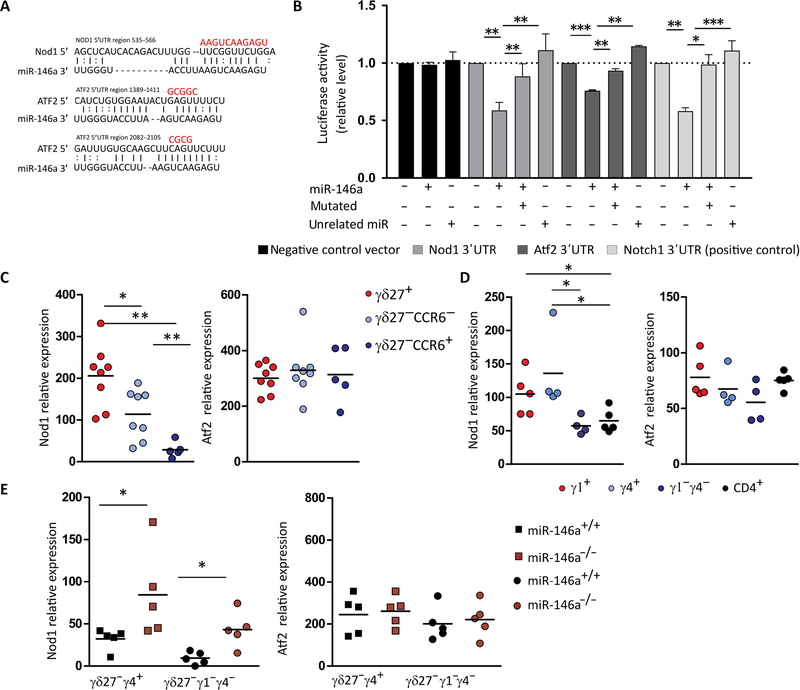
Fig. 5. miR-146a targets Nod1 mRNA that is depleted in γδ27− T cells.
(A) Putative binding sites of miR-146a-5p in the 3′UTR region of Nod1 and Atf2. The line represents canonical Watson-Crick base pairing, whereas the dot represents noncanonical base pairing between miR-146a and Nod1 or Atf2 mRNAs. The red bases above illustrated the mutated sites in the region of Nod1 and Atf2 3′UTRs. (B) Dual luciferase reporter assay was performed to verify binding between miR-146a and Nod1 or Atf2 mRNAs. HEK293 T cells were cotransfected with a pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega) containing either the WT or mutated 3′UTR target sites plus miR-146a, an unrelated miRNA, or GFP control expression vector. A negative construct (without miR-146a binding sites) and a positive construct (3′UTR of Notch1) were included. Data are from three to four independent experiments with technical replicates. *P < 0.05, **P < 0.005, ***P < 0.001, Student’s t test. RT-qPCR analysis of Nod1 and Atf2 and expression in (C) sorted γδ27+, γδ27−CCR6−, and γδ27−CCR6+ T cells from pooled peripheral organs (lymph node and spleen) of C57BL/6 mice; (D) sorted Vγ1+, Vγ4+, and Vγ1−γ4− γδ T cells and CD4+ T cells from pooled peripheral organs (lymph node and spleen) of C57BL/6 mice; and (E) sorted Vγ4+CD27− and Vγ1−γ4− CD27-γδ T cells from pooled lymph nodes of either miR-146a+/+ or miR-146a−/− mice. Results are presented relative to β-actin and HPRT expression. Each symbol in (C) to (E) represents an individual mouse. *P < 0.05 and **P < 0.01 (Mann-Whitney two-tailed test).