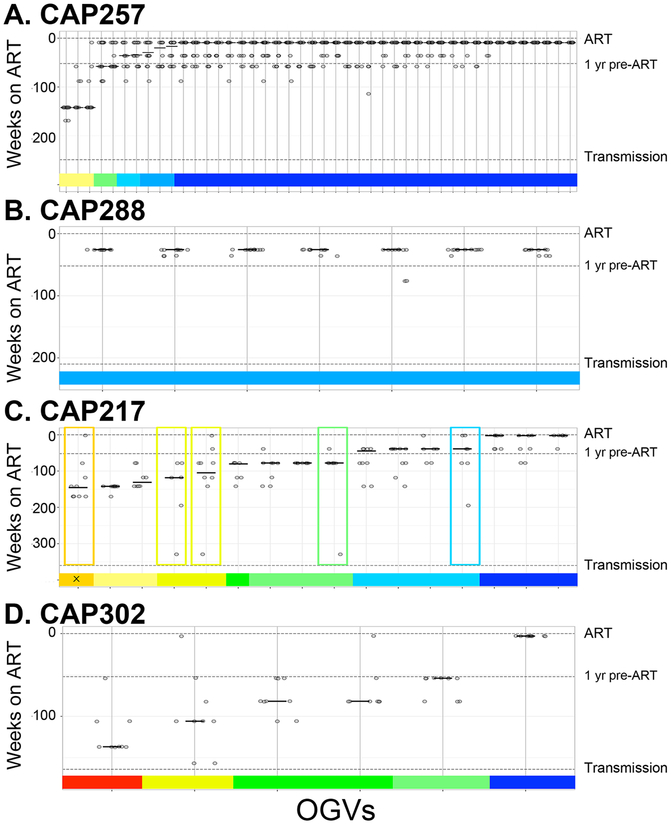
Fig. 4. Timing of Reservoir Entry for OGVs as Estimated by Combining Three Methods.
Three methods (patristic distance, clade support and phylogenetic placement) were used to generate estimates of OGV entry into the reservoir by comparing multiple regions of the OGV genome to that of viral RNA isolated from the plasma pre-ART. (a, b) Estimates are for two participants with OGVs that typically entered the reservoir near the time of ART initiation (CAP257 and CAP288) and (c, d) two with OGVs from throughout untreated infection (CAP217 and CAP302). Each OGV is represented as a vertical line with up to 15 estimates along that line. For each OGV, estimates of reservoir entry were summarized as a weighted median, shown as a horizonal line and represented in a colored bar along the x axis. Boxes indicate OGVs with highly variable estimates (SD > 1 yr). Additional phylogenetic analyses of OGVs with variable estimates found that one (designated with an ‘x’) was in a time-specific lineage that did not correspond to the weighted median. All other OGVs were found in time-specific lineages that corresponded to the weighted median.