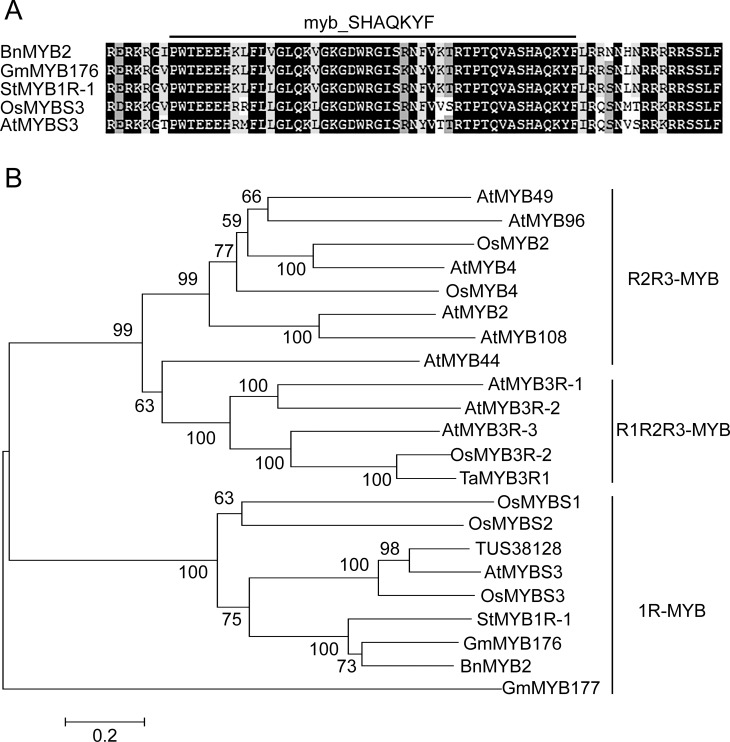
Fig 1. Multiple alignment and phylogenetic analysis of BnMYB2 and MYB proteins related to abiotic stress from other plants.
A, The alignment of BnMYB2 and its ortholog proteins of different plants constructed using DNAMAN 8.0 software. Black shadow represents the conserved residues. The line indicates a highly conserved MYB DNA-binding domain (myb_SHAQKYF). B, Neighbor-joining phylogenetic tree of BnMYB2 from Boehmeria nivea and other MYBs constructed using MEGA 5.0 software. The scale bar represents 0.05 amino acid substitutions per site. The statistical reliability of individual nodes of the tree is assessed by bootstrap analyses with 1 000 replications. GenBank accession numbers of the proteins are as follows: AtMYB49 (At5g54230), AtMYB96 (At5g62470), AtMYB4 (At4g38620), AtMYB2 (At2g47190), AtMYB108 (At3g06490), AtMYB44 (At5g67300), AtMYB3R-1 (At4G32730), AtMYB3R-2 (At4g00540), AtMYB3R-3 (At3g09370) and AtMYBS3 (At5g47390) from Arabidopsis thaliana; OsMYB2 (BAA23338), OsMYB4 (BAA23340), OsMYB3R-2 (BAD81765.1), OsMYBS1 (AAN63152.1), OsMYBS2 (AAN63153.1) and OsMYBS3 (AAN63154.1) from Oryza sativa; TaMYB3R1 (ADO32617.1) from Triticum aestivum; TUS38128 (XP_004508939.1) from Cicer arietinum; StMYB1R-1 (ABB86258.1) from Solanum tuberosum; GmMYB176 (NP_001236048.2) and GmMYB177 (ABH02866.1) from Glycine max.