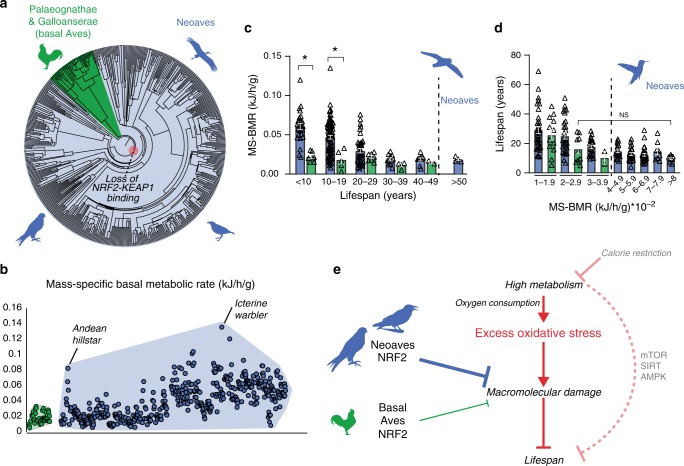
Fig. 5. Diversification of lifespan and metabolic rates in Neoaves.
a Phylogenetic relationships of avian taxa for which mass-specific basal metabolic rates (MS-BMR) are available (“Methods”). Red highlight denotes the Neoavian ancestor where KEAP1 binding of NRF2 was lost. b MS-BMR reach higher ranges in Neoaves (blue) species relative to basal Aves (green). Avian orders are grouped by phylogenetic relationships along the X-axis (“Methods”). c MS-BMR differs significantly between Neoaves and basal Aves species with equivalent lifespans (<10 years, p = 0.03; 10–19 years, p = 0.02; PI Kruskal–Wallis; Supplementary Fig. 9, Supplementary Tables 10 and 11). Neoaves with low MS-BMR reach higher lifespans than any basal Aves (>50 years). Neoaves: n = 180 biologically independent animals; basal Aves: n = 25 biologically independent animals. Data are presented as mean values. d Neoaves with MS-BMR values higher than that of any basal Aves still maintain statistically indistinguishable lifespans, despite a nearly fourfold increase in MS-BMR (Kruskal–Wallis; Supplementary Table 10). Neoaves: n = 175 biologically independent animals; basal Aves: n = 23 biologically independent animals. Data are presented as mean values. e High metabolism impacts lifespan through nutrient-sensing pathways (mTOR, SIRT, AMPK) as well by generating excess oxidative stress, aggravating age-associated cellular and molecular damage (see “Discussion” for references). An enhanced NRF2 antioxidant response via the loss of KEAP1 would likely lower the risk of macromolecular oxidative damage expected to result from high avian metabolism and oxygen consumption. By altering redox homeostasis to tolerate higher levels of excess oxidative stress, the loss of KEAP1 may have enabled these increases to Neoaves lifespan and metabolic rates. Source data are provided as a Source data file. All error bars represent standard error. * = phylogenetic statistical significance.