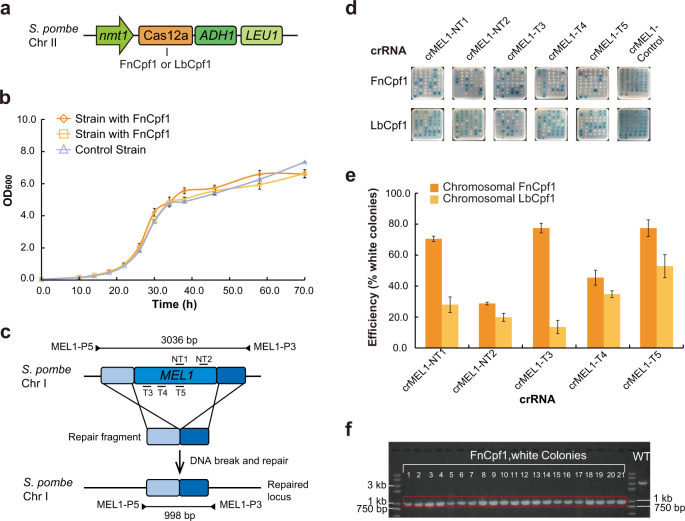
Fig. 1. Implementation of Cas12a editing system in S. pombe and editing of MEL1 gene.
a Schematic representation of the S. pombe chromosomal locus with integrated Cas12a constructs. b Growth in liquid culture media (EMM + uracil) of S. pombe strains expressing FnCpf1 or LbCpf1, compared to the control strain with blank insertion at the same locus. They had an average double time of 4.1, 4.6 and 4.5 h, respectively. Data are means ± SEM (n = 4 independent experiments). c Schematic representation of MEL1 gene editing. crRNAs were designed to target five sites (NT1, NT2, T3, T4, and T5) within MEL1 gene. The DNA cut by Cas12a is repaired with the repair fragment by HR. The arrows denote the primers, MEL1-P5 and MEL1-P3, and their PCR products for diagnostic experiments. d The editing efficiency on MEL1 gene with five different crRNAs, estimated by formation of white- and blue-colored colonies, assayed on agar plates containing the X-α-Gal. e Efficiency of genome editing on MEL1 gene with five different crRNAs. Values are mean ± SEM, n = 3. f Diagnostic PCR products of mel1 mutants (white colonies) generated from FnCpf1 editing, showing 998-bp in size. They are compared with the wild-type (WT) product, 3036-bp in size. Primer sets MEL1-P5 and MEL1-P3 (c) were used for PCR.