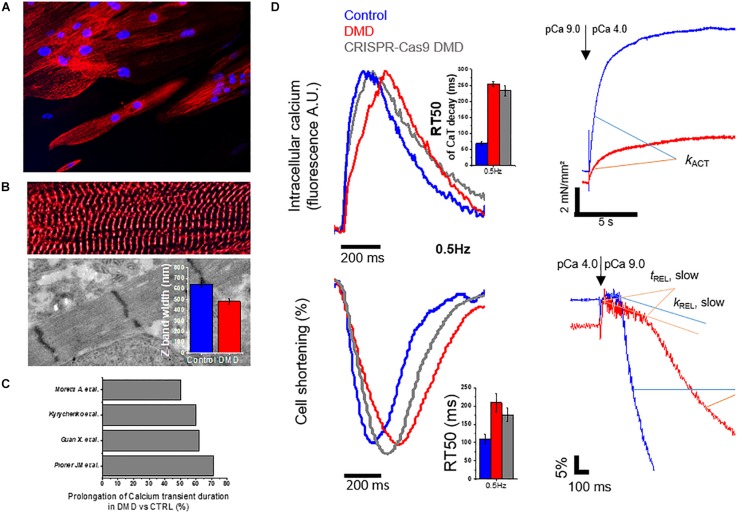
FIGURE 4.
Morphology and function altered in DMD-hiPSC-CMs from patient confirmed in a CRISPR-Cas9 gene edited cell line. Original data are modified from Pioner et al. (2019b) with the correct permission from the owner (Pioner et al., 2019a). A DMD-hiPSC-cell line from a patient (with Δ Exon 50 in DMD gene) and a CRISPR-Cas9 gene edited cell line (c.263 ΔG) targeting Exon 1 in healthy control cell line (Control) were generated and the cardiomyocytes were matured onto nanotopographic cues for 3 months. (A) DMD-hiPSC-CMs displayed aspect ratio similar to adult cardiomyocytes and Z-bands were observed across the entire cell width (diameter), suggesting DMD-CMs experienced hypertrophy and a greater number of myofibrils in parallel. (B) Despite similar myofibril alignment to control-hiPSC-CMs, sarcomere diameters (estimated from the length of Z-bands by transmitted electron microscopy) were significantly smaller, suggesting a possible reduction in the parallel assembly of myofilaments within individual myofibrils. (C) Meta-analysis on Calcium transient duration. (D) Representative traces of calcium transients (CaT), cell shortening and myofibril mechanics of DMD-hiPSC-CMs compared to controls. Simultaneous recordings at single cell level revealed slower CaT decay (estimated from time to peak to 50% of CaT decay, RT50, ms), slower cell relaxation (RT50, ms) at 37°C and external pacing (0.5 Hz reported). Compared to other studies reporting similar analysis of calcium transients, slower CaT duration might be a peculiarity of DMD-hiPSC-CMs. Single myofibrils showed lower isometric-tension generating capacity and slower myofibril relaxation (slow tREL and fast kREL). This study concluded that both calcium handling and myofibril abnormalities may contribute to prolong cell relaxation.