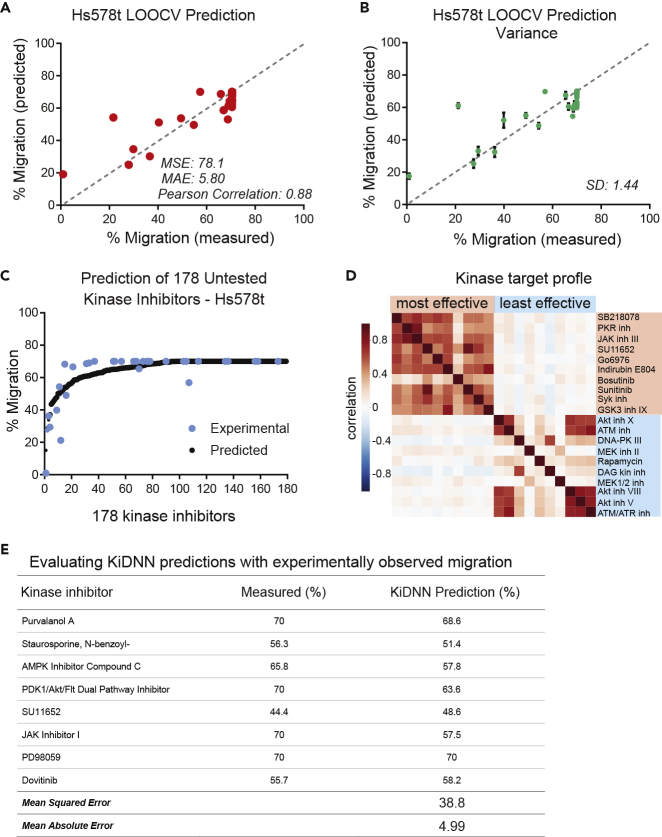
Figure 4.
Predicting Response to Naive Kinase Inhibitors
(A) A plot showing comparison of KiDNN-predicted and measured percent migration in response to 32 inhibitors in Hs578t cells using LOOCV. Migration in response to untreated/DMSO control was 70%. The MSE, MAE, and Pearson correlations are also indicated.
(B) KiDNN model was run for 10 iterations to generate predictions for all 32 inhibitors. Green circles denote the mean predicted migration, and the error bars show the standard deviation of predictions for each kinase inhibitor. The standard deviation is also listed.
(C) A plot showing KiDNN-predicted response to 178 small molecule kinase inhibitors (146 untested). The black circles denote the predicted percent migration, whereas blue circles denote the 32 experimentally validated kinase inhibitors. The kinase inhibitors are ranked by predicted migration.
(D) A heatmap showing correlation of kinase target profiles of the 10 most and 10 least effective inhibitors predicted by KiDNN.
(E) A table showing comparison of KiDNN-prediction and experimental changes in migration of Hs578t cells in response to eight kinase inhibitors. Each inhibitor was tested at multiple doses (10 nM–10 μM), and the effect of kinase inhibitor at 500 nM dose was calculated using the dose-response curves. Hs578t cells treated with DMSO control showed migration of 70%.