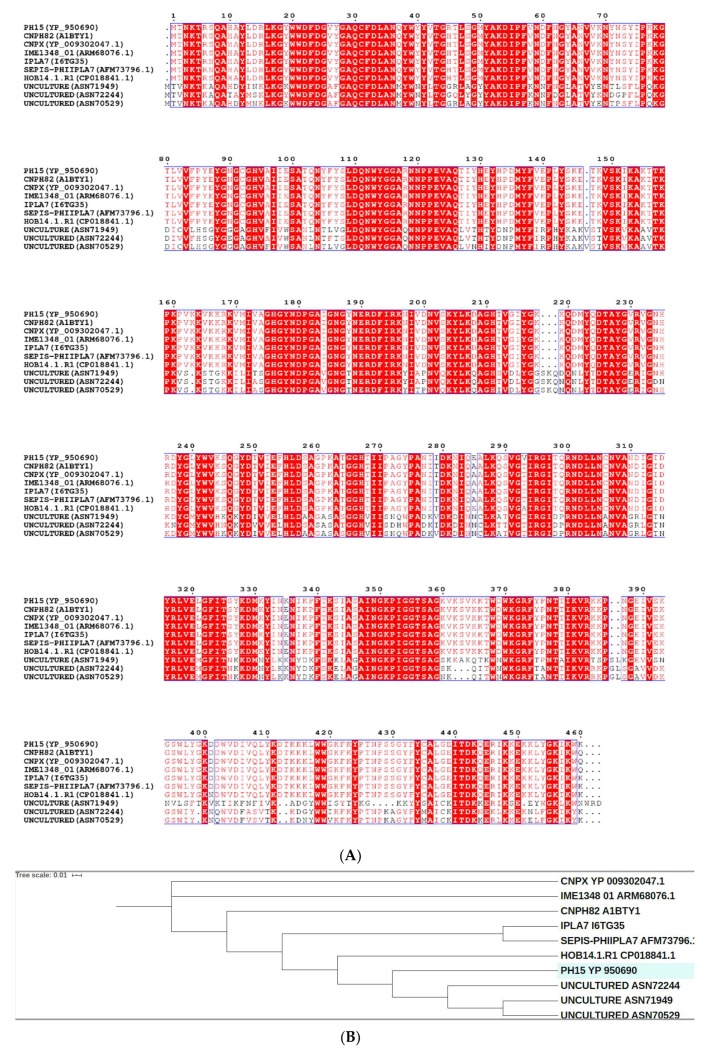
Figure 3.
(A). Amino acid sequence alignements of endolysin Ph28 with other putative endolysin sequences from Staphylococcus viruses that infect S. epidermidis strains. The alignments were produced using Clustal Omega [26]. Conserved areas are shown shaded. A column is framed, if more than 70% of its residues are similar according to physico-chemical properties. (B). Phylogenetic analysis of endolysin Ph28. Phylogenetic tree was constructed by Neighbour-Joining method using the iTOL programme [27]. The tree was formed after alignment of the protein sequences using Clustal Omega [26]. The endolysin sequences were derived from: Staphylococcus virus CNPH82 (NCBI accession number A1BTY1), Staphylococcus virus IPLA7 (NCBI accession number I6TG35), Staphylococcus phage CNPx (NCBI accession number YP_009302047.1), Staphylococcus phage vB_SepiS-phiIPLA7 (AFM73796.1), Staphylococcus phage IME1348_(NCBI accession number ARM68076.1), uncultured Caudovirales phage (NCBI accession number ASN72244), uncultured Caudovirales phage (NCBI accession number ASN70529), uncultured Caudovirales phage (NCBI accession number ASN71949), Staphylococcus phage HOB 14.1.R1 (NCBI accession number CP018841.1).