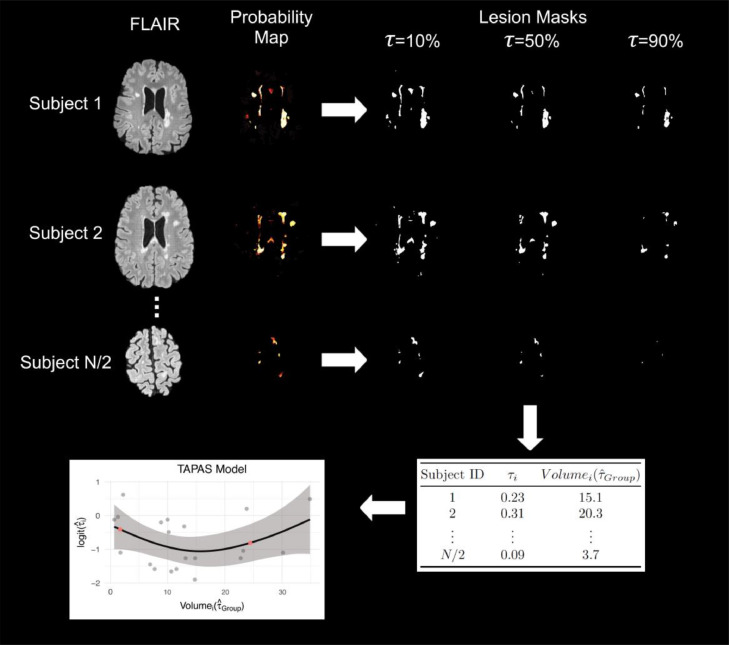
Figure 2.
The TAPAS procedure is shown using sample axial slices from the data. A set of training scans with manual delineations were used to train and apply MIMoSA in order to obtain probability maps. For each subject's probability map, we applied thresholds at to 100% by 1% to create estimated lesion masks. For simplicity, in this example, we have only shown , 50%, and 90%. Based on Sørensen-Dice similarity coefficient (DSC) calculations within and across subjects we estimated and . Using we obtained . We fit the TAPAS model and applied it to subjects in the test set to determine . Red points in the plot represent and , or lower and upper bounds at the volume associated with the 10th and 90th percentiles, respectively.