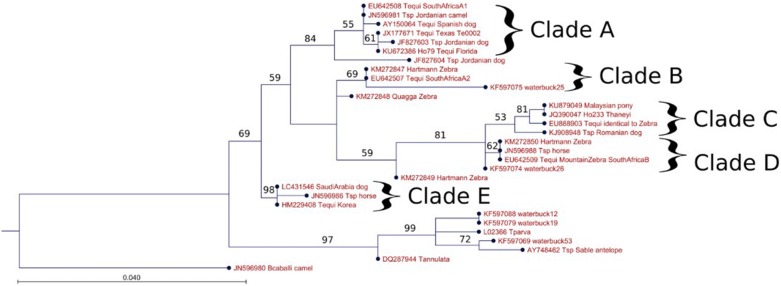
Fig. 1.
18S rDNA sequences placed in GenBank and identified as Theileria equi but detected within various host species were gathered and aligned in CLC Genomics Workbench v.10.1.1 (Qiagen), along with outgroup 18S rDNA sequences from other piroplasmids as identified in the figure. All sequences were trimmed to identical length of 401 bp, and span the 4th hypervariable region of full-length 18S rDNA. Aligned sequences were compared in a maximum likelihood phylogeny in the same program, with a Kimura 80 nucleotide substitution model, with neighbor-joining used in constructing initial trees to enable enhanced evolutionary inference. Five hundred replicates were performed in a bootstrap analysis with the values for nodes above a cut-off of 50 indicated on the tree for major branches. The tree provides a representative sample of ribosomal sequences falling into distinct clades across different infected host species and is not is intended to provide an exhaustive depiction of the relationships of all equid Theileria isolates