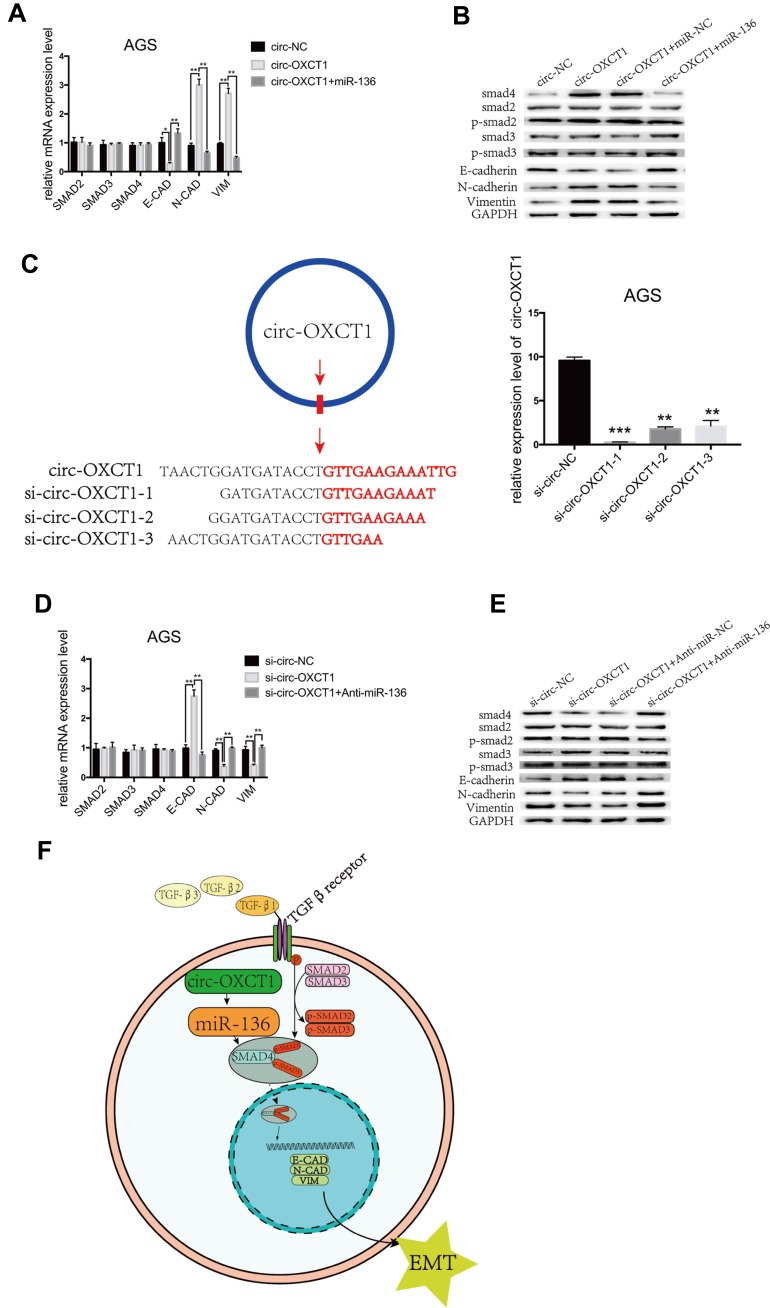
Figure 6.
Circ-OXCT1 suppressed SMAD4 expression and EMT via the circ-OXCT1/miR-136/SMAD4 axis in GC cells. (A, B) mRNA levels of SMAD2, SMAD3, SMAD4, E-CAD, N-CAD, VIM and protein levels of SMAD2, p-SMAD2, SMAD3, p-SMAD3, SMAD4, E-CAD, N-CAD, VIM are shown after cotransfected with circ-OXCT1 and miR-136 mimics in AGS (*p < 0.05, **p < 0.01). (C) Schematic display of the targeting sequences of siRNAs (si-circ-OXCT1-1, si-circ-OXCT1-2 and si-circ-OXCT1-3) specific to the back-splice junction of circ-OXCT1. Si-circ-OXCT1-1 was verified the most effectively inhibitory siRNA (red dart represent the back-splice junction) (**p < 0.01, ***p=0.00). (D, E) Similar results were observed when cotransfected with si-circ-OXCT1-1 and anti-miR-136 (**p < 0.01). (F) A proposed mechanistic model in which circ-OXCT1 functions as a miR-136 sponge and regulates SMAD4/TGF-β-induced EMT in GC cells. GAPDH was used as a loading control.
Abbreviations: E-CAD, E-cadherin; N-CAD, N-cadherin; VIM, vimentin.