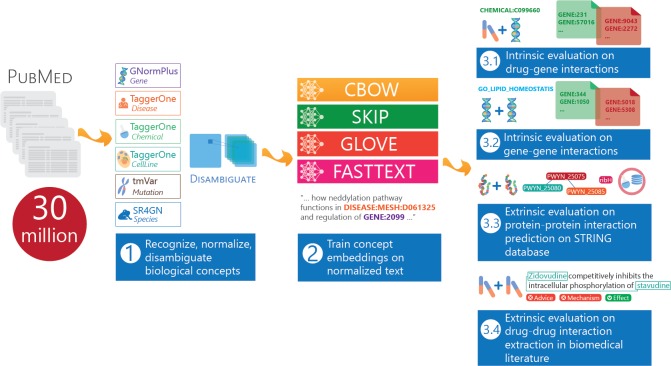
Fig 1. An overview of our study.
BioConceptVec was trained on PubMed abstracts, which consists of ~30 million documents. (1) We employed PubTator, which contains four NER tools, to annotate and normalize the concepts. (2) We trained four concept embeddings on the normalized corpus. (3) We conducted both intrinsic evaluations on drug-gene interactions and gene-gene interactions, and extrinsic evaluations on protein-protein interaction prediction and drug-drug interaction extraction to evaluate the effectiveness of BioConceptVec.