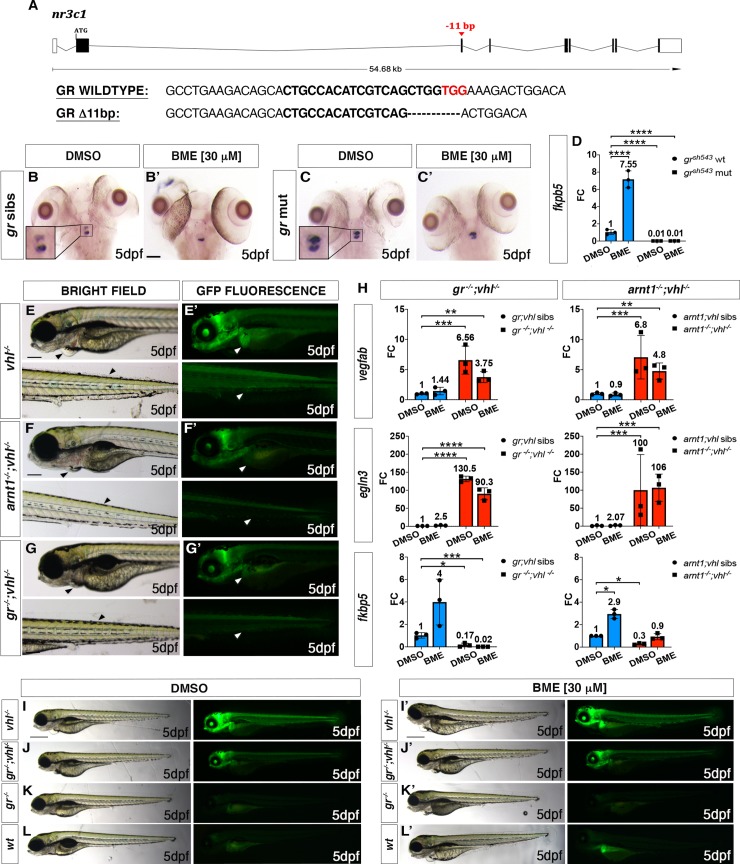
Fig 3. gr mutation partially rescues vhl phenotype.
A. Schematic representation of zebrafish gr (nr3c1) gene. Exons are shown as boxes, introns as lines. The red arrowhead shows the position of a -11 bp deletion in exon 3 (encoding the DNA binding domain). gr wt and mutant sequence. CRISPR target site: bold. PAM sequence: red. B-C’. Representative pictures of WISH performed on DMSO and BME [30 μM] treated gr mutant line, at 5 dpf, using pomca as probe. Scale bar 100 μm. gr siblings DMSO treated (n = 30/30 larvae) showed normal expression; gr siblings (n = 29/30 larvae) showed downregulated pomca expression after BME treatment. Both DMSO treated (n = 30/30) and BME treated (n = 30/30) gr-/- larvae showed upregulated pomca expression. D. RTqPCR analysis performed on gr wt (n = 10; 3 repeats) and gr-/- (n = 10; 3 repeats) larvae at 5 dpf, using fkbp5 as probe. Statistical analysis was performed on ΔΔCt values, whereas data are shown as fold change values. Ordinary Two-way ANOVA followed by Dunnett’s multiple comparison test (****P < 0.0001). E-G. Magnified picture of representative gr-/-; vhl-/- larvae compared to arnt1-/-;vhl-/- and vhl-/- larvae. Both double mutants are characterized by the absence of pericardial oedema, no ectopic extra vasculature at the level of the tail, no bright liver and a reduced brightness in the rest of the body (white and black arrowheads), compared to vhl-/- larvae. Fluorescence, exposure = 2 seconds. Scale bar 200 μm. H. RTqPCR analysis performed both on HIF and GC target genes expression carried out on gr-/-; vhl-/- and sibling at 5 dpf, (n = 10 larvae, per group, in triplicate) compared to arnt1-/-;vhl-/- larvae and siblings, at 5dpd (n = 10 larvae, per group, in triplicate). Both vegfab and egln3 are HIF target genes, whereas fkbp5 is a GC target gene. Statistical analysis was performed on ΔΔCt values, whereas data are shown as fold change values, Ordinary Two-way ANOVA followed by Dunnett’s multiple comparison test. I-L. Representative picture of phenotypic analysis performed on DMSO and BME [30 μM] treated gr+/-; vhl+/-(phd3:eGFP) incross-derived 5 dpf larvae (n = 600). All the genotype combinations observed are represented in the figure. Among the 450 GFP+ larvae analysed, 28 showed a partially rescued vhl phenotype which resembled the arnt1’s one. Three experimental repeats. In all panels: *P < 0.05; **P < 0.01; ***P <0.001; ****P < 0.0001. Fluorescence, exposure = 2 seconds. Scale bar 500 μm.