Figure 4.
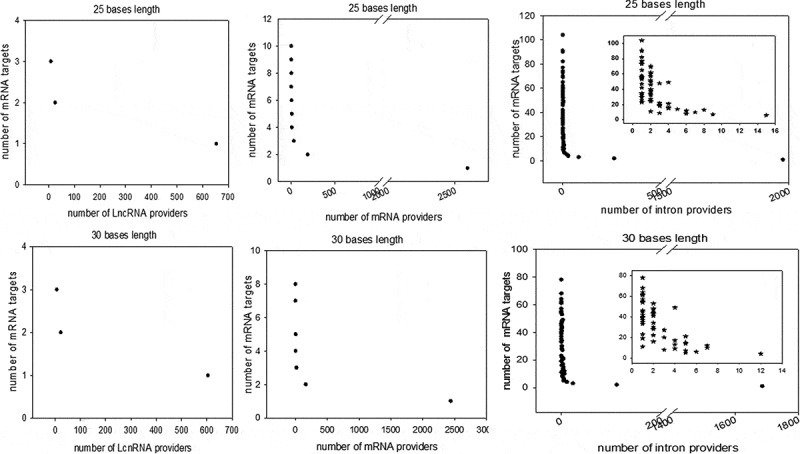
Distribution of computationally identified dsRNAs considering minimum perfect base pairing lengths of 25 and 30 bases. The duplex RNAs were analysed as the number of targets (mRNAs) by provider and the number of providers (LncRNA, mRNA and introns) by target (mRNA). For each of our computationally generated lists, we counted the number of individual target mRNAs that matched 1 to x different providers; the figure shows the series for lncRNAs (left), mRNAs (centre) and introns (right). Counts corresponding to exact matches of at least 25 and 30 bases are presented at the top and bottom, respectively. A high-magnification zoom showing the details of the distribution of the lowest values is presented for the intron/mRNA analysis.
