Figure 9.
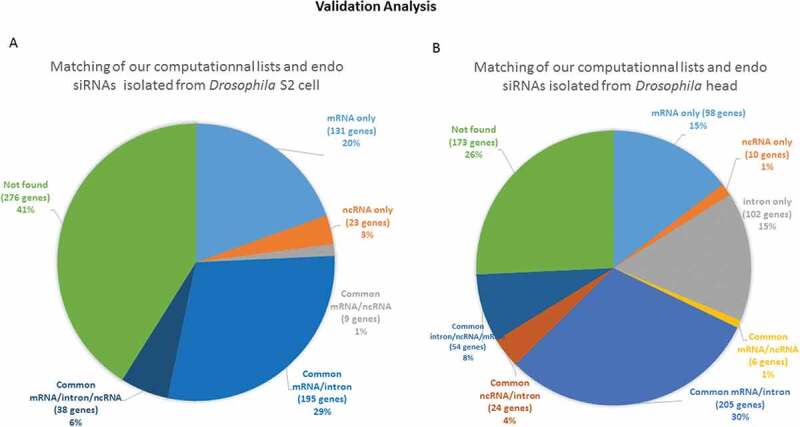
Validation of our computationally generated lists by comparison with a published sequence dataset of isolated endo-siRNAs A comparison of the list of sequences corresponding to the isolated endo-siRNAs reported by Ghildiyal et al. and our computationally derived list of endo-siRNAs was performed. The circular diagrams show the isolated and sequenced species and those that were also found in our computational lists. The percentages of the terms associated with the genes from which the endo-siRNAs are derived are presented. (A) The distribution of genes in the sequenced species list retrieved from Drosophila S2 cells that were found in our mRNA target/provider lists; (B) the distribution of genes related to the list of RNAs retrieved from Drosophila head tissue and found in our lists. The segments marked ‘not found’ were isolated and sequenced fragments that did not conform to the terms of our computationally established lists. ‘Only’ means that the isolated sequences were found in only one computational list; ‘common’ means that the relevant isolated sequences were found in at least two computational lists.
