Figure 4.
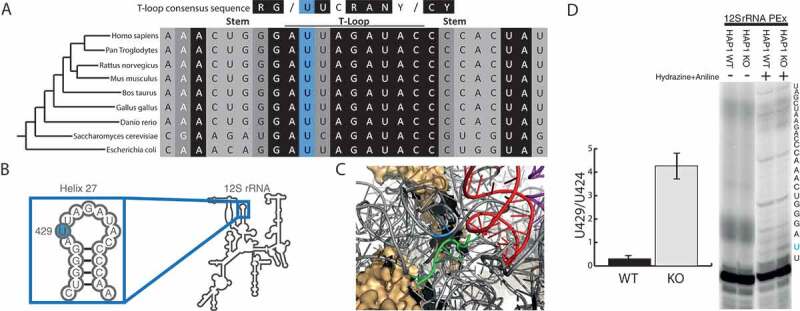
TRMT2B catalyses m5U429 in 12S mitochondrial rRNA.
(A) Alignment of the small ribosomal RNA (rRNA) from a range of species in a region corresponding to helix 27 in human 12S mt-rRNA. The degree of shadowing represents the extent of conservation for a given residue, with U429 (or the corresponding position in other species) shown in blue background. The T-loop consensus sequence is displayed above for comparison. (B) Schematic of human 12S mt-rRNA, helix 27, and the location of m5U429 (blue circle). (C) The structure of the human mitoribosome highlighting U429 (blue), the bound mRNA (green), and the adjacently bound tRNA (red). (D) Separation and detection of RT-PEx products using a [32P]-end labelled primer complementary to the region upstream of m5U429 in 12S rRNA. Extension reactions performed on RNA derived from a HAP1 wild-type (WT) and a HAP1 TRMT2B knockout cell line (KO), with or without hydrazine-aniline treatment. The nucleotide sequence of 12S rRNA is shown to the side of the panel. Quantification values represent the ratio between stalling at U429 and stalling at the next uridine residue (U424). Error bars = SEM, n = 4.
