Figure 5.
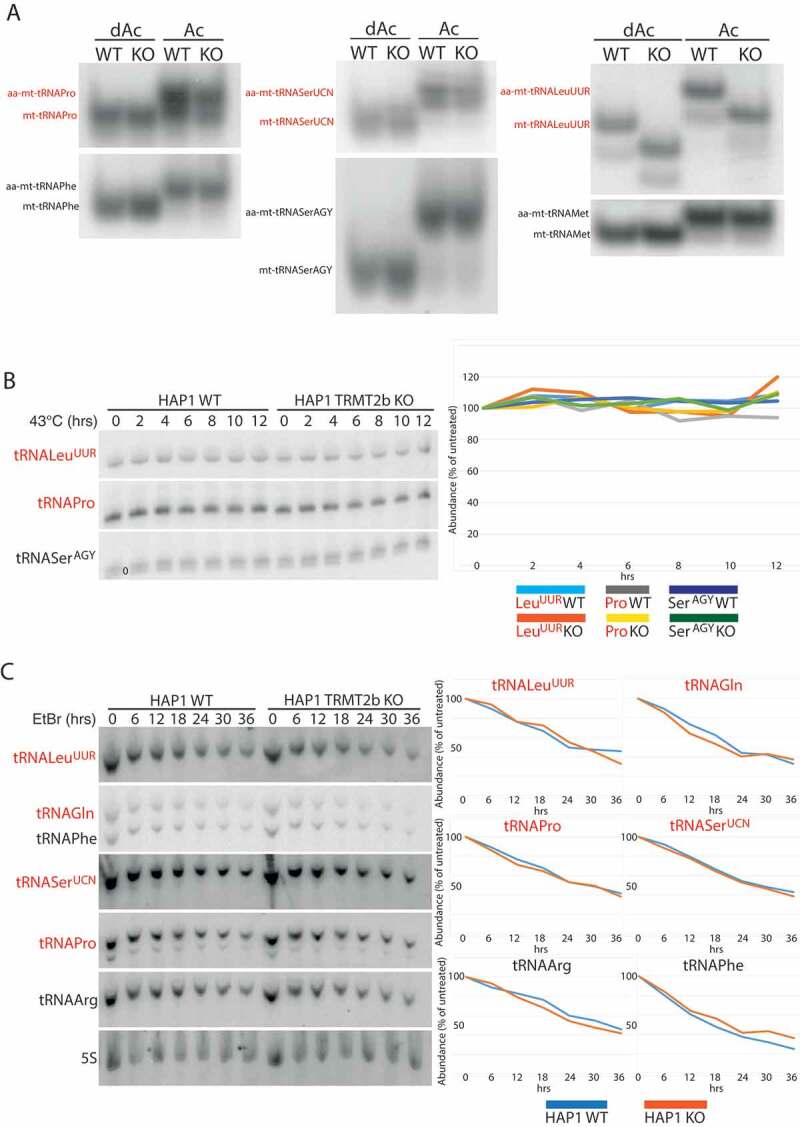
tRNA aminoacylation and stability following the loss of TRMT2B.
(A) High resolution acidic polyacylamide gel electrophoresis (PAGE) northern blot analysis of total RNA extracted from HAP1 wild-type (WT) or TRMT2B knockout cell line (KO). RNA was either maintained in low pH conditions to preserve aminoacylation (Ac), or intentionally deacylated prior to being run on the gel (dAc). The blots were probed with the mt-tRNA-specific riboprobes as indicated, with the tRNAs known to contain m5U54 in red text.(B) High resolution PAGE northern blot analysis of total RNA extracted from HAP1 parental cell line (WT), or a HAP1 TRMT2B knockout cell line (KO), following their growth at 43°C for between 2 and 12 hours. Control cells kept at 37°C shown as ‘0 hrs’. The blots were probed with the mt-tRNA-specific riboprobes as indicated, with the tRNAs known to contain m5U54 in red text. Quantification values were plotted as a percentage of the untreated 0 hr RNA. (C) As above following exposure to 250 μg/mL ethidium bromide (EtBr) for between 6 and 36 hours. Control cells grown in media containing no EtBr shown as ‘0 hrs’. The blots were probed with the mt-tRNA-specific riboprobes as indicated, with the tRNAs known to contain m5U54 in red text. Quantification values were plotted as a percentage of the untreated 0 hr RNA, after they had been normalized to 5S to control for RNA loading.
