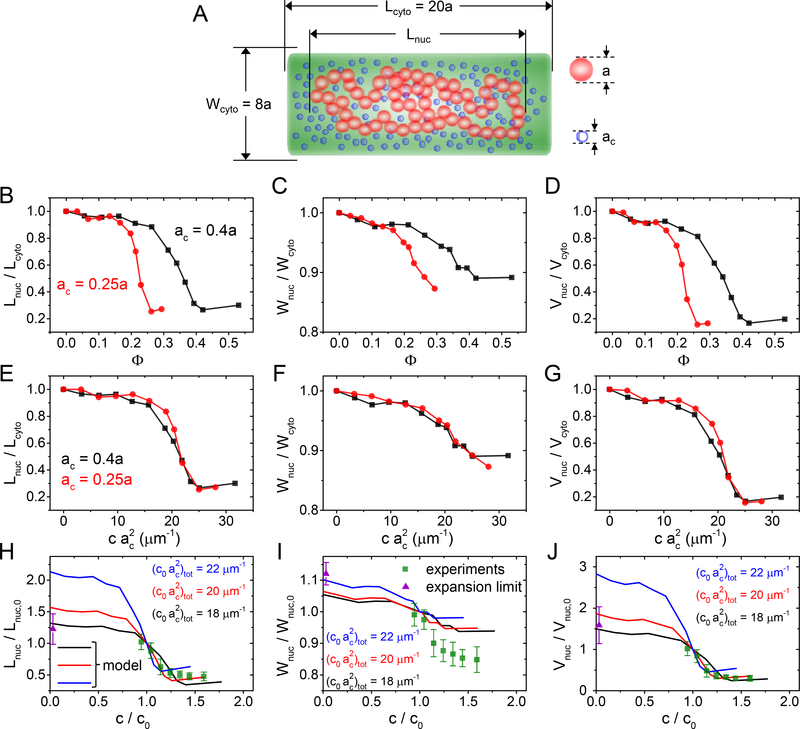
Figure 6. Comparing a polymer model to the experimental data.
(A) A model of DNA consists of 150 beads with diameter a that are confined to a cylindrical volume. The same volume is also occupied by a variable number of crowders having diameters ac. For details of the model see SI Text: Coarse-grained Brownian Dynamics simulations. (B-D) Calculated nucleoid length, width, and volume for two different size crowders as a function of their volume fraction. (E-G) Calculated nucleoid length, width, and volume as a function of effective “crowding level” defined as . The curves with different crowder diameters collapse into a single one. (H-J) Comparing modelled nucleoid length, width, and volume (solid lines) to experimental data from slow growth conditions (green squares; from Fig. 2G–F). The experimental data is augmented by a data point at the zero crowder concentration limit where the nucleoid is expected to extend over the whole cytosolic volume (violet triangle). A set of curves from the model with “crowding level” 18, 20, 22 μm−1 are plotted. Of those the best agreement between the experiment and the model is at 20 μm−1. The error bars for the data are std.